FIGURE 1.
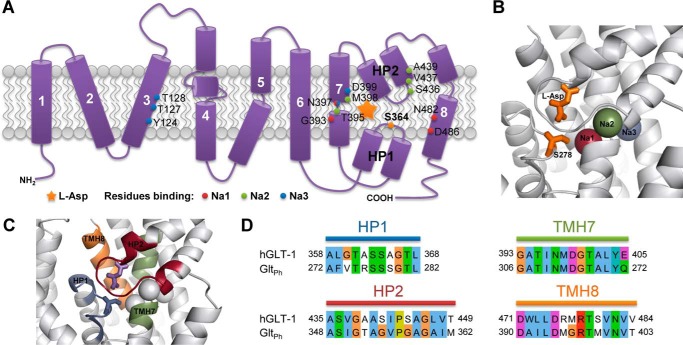
Amino acid residues involved in substrate and sodium ion binding of the glutamate transporter SLC1A2 and its archaeal homologue GltPh. A, schematic model of human SLC1A2 topology, representing the eight transmembrane helices and hairpin loops HP1 and HP2. The location of the bound substrate is marked by a star. Residues involved in sodium ion binding according to GltPh x-ray structures are indicated and color-coded. Note that Asn-397 is binding both Na1 and Na3. The herein studied Ser-364 residue is marked in orange within HP1. B, structure of the substrate binding site in GltPh (PDB ID: 2NWX). Sodium ions, bound l-aspartate substrate, and Ser-278 (corresponding to Ser-364 in human SLC1A2) are shown and color-coded according to panel A. The location of Na3 is taken from Bastug et al. (29). C, protein regions considered to be in the neighborhood of the substrate binding site are highlighted in color. D, conservation of the substrate binding site and its neighborhood as shown on panel C between human SLC1A2 (hGLT-1) and GltPh. Sequence identity of the selected regions is 64.2%, as opposed to 36% for the full-length proteins.