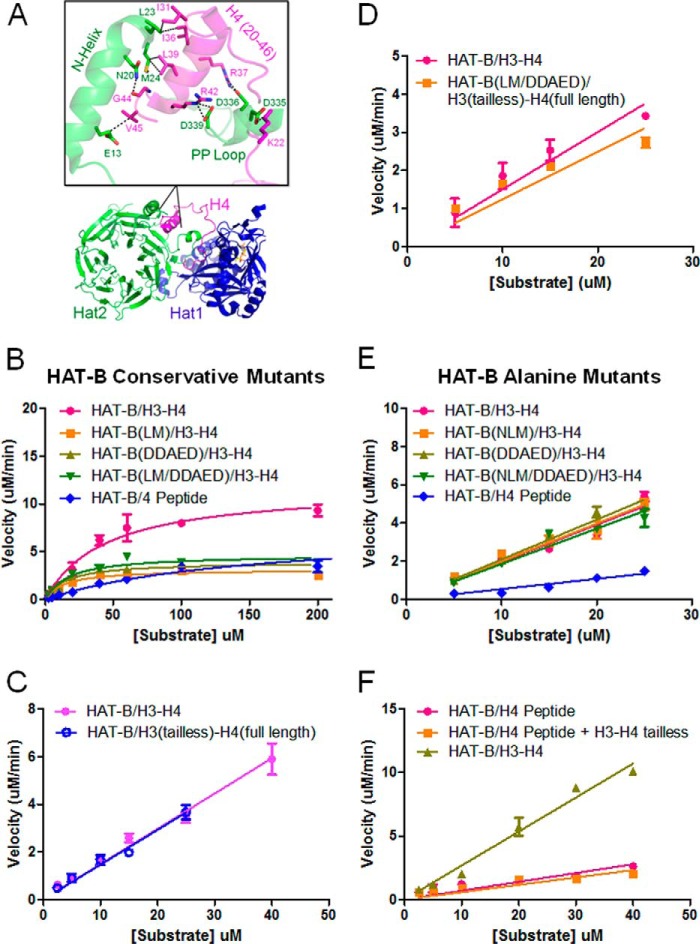
FIGURE 5.
Mutational analysis of HAT-B acetylation of the H3-H4 heterodimer. A, crystal structure of HAT-B bound to H4 (32) with the binding region of Hat2 and H4 enlarged to highlight key interacting residues. B, enzyme kinetics of Hat2 structurally conservative mutations. HAT-B and HAT-B containing Hat2 mutants with saturating levels of 14C-labeled acetyl CoA and varying concentrations of either H3-H4 (1–75 μm) or H4 peptide (2.5–1000 μm) are fit to Michaelis-Menten kinetics (GraphPad Prism 5) to calculate steady state parameters. C, enzyme kinetics of HAT-B with saturating concentrations of 14C-labeled acetyl CoA with either H3-H4 (pink) or tail-less H3(45–135)-H4 (H3tl-H4) (blue) substrates (0.5–25 μm) are fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. D, enzyme kinetics of HAT-B with either H3-H4 compared with Hat2 mutants on tail-less H3tl-H4 substrates as described for C. E, enzyme kinetics of Hat2 structurally disruptive mutations as described for C. F, enzyme kinetics of wild-type HAT-B varying both H4 peptide added in trans with tail-less H3(45–135)-H4(20–102) (H3tl-H4tl; 0.5–40 μm) is fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. All reactions were performed using 100 nm enzyme at saturating concentrations of 14C-labeled acetyl CoA (500 μm), and all kinetic measurements were carried out in duplicate.