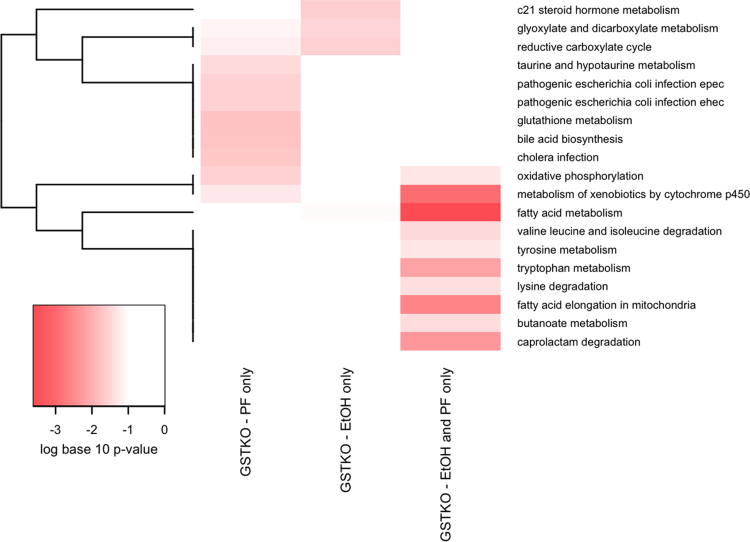
Fig. 5.
KEGG Bioinformatic analysis of mitochondrial carbonylated proteins unique to GSTA4−/− PF, GSTA4−/− EtOH or PF/EtOH groups. For enrichment analysis, three groups of proteins were identified and enrichment of pathways for each group are represented in each column. The GSTKO-PF only column represents pathways enriched for proteins identified in at least 50% of the pair-fed GSTA4−/− mice, but less than 50% of samples in all three of the other groups and not identified in the background sample. The GSTKO-EtOH only column represents pathways enriched for proteins identified in at least 50% of the ethanol-fed GSTA4−/− mice, but less than 50% of samples in all three of the other groups and not identified in the background sample. The GSTKO-EtOH and PF only column represents pathways enriched for proteins identified in at least 50% of the ethanol-fed GSTA4−/− mice and at least 50% of pair-fed GSTA4−/− mice, but less than 50% of samples in the SV groups and not identified in the background sample. Proteins functionally annotated using KEGG pathways [23] were examined for enrichment using EnrichR [24]. KEGG pathways that were nominally significant (p<0.05) in at least one of the 3 protein lists are included in the graphic. The colors of the heatmap range from white (unadjusted p-Value>0.05) to bright red based on the log base 10 transformation of the unadjusted p-value. A p-value of 1 was used when the KEGG pathway was not represented by any proteins in the list. KEGG pathways (rows) are ordered based on hierarchical clustering using the Euclidean distance and a binary indicator of significance (p<0.05 vs. p>=0.05).