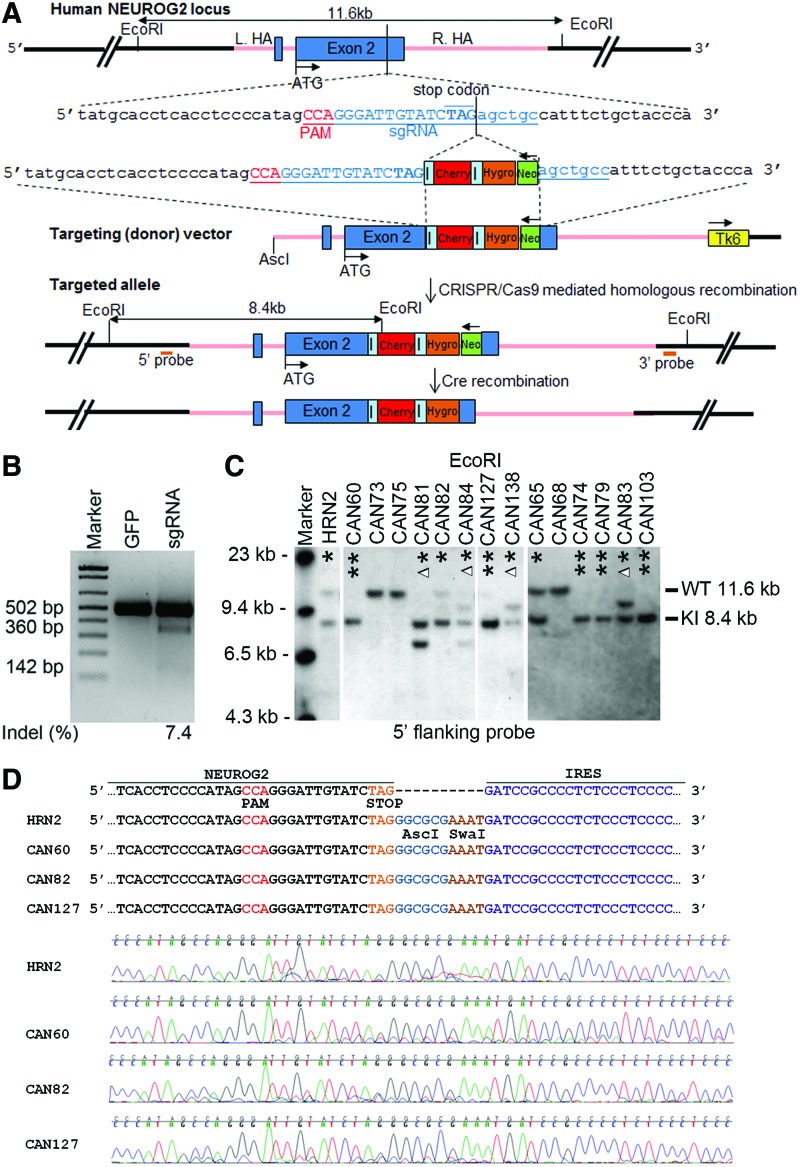
FIG. 1.
Gene targeting to the human neurogenin2 (NEUROG2) locus to create dual reporter human induced pluripotent stem cell (hiPSC) lines. (A) Design of targeting vector and single guide RNA (sgRNA) for making knockin NEUROG2-IRES-mCherry-IRES-hygromycin dual reporter hiPSC lines. The NEUROG2 genomic sequence, which consists of two exons (two blue boxes) and one intron, is tagged by mCherry (red box) and hygromycin resistance cassette (orange box) at the end of exon 2, connected in tandem by two internal ribosome entry site (IRES) sequences (light blue bar). The endogenous NEUROG2 gene is intact and NEUROG2 and the two tags mCherry and hygromycin resistance gene are driven by the endogenous NEUROG2 promoter. A floxed neomycin cassette (green box, driven by RNA pol II promoter) is used for positive selection. Tk6 cassette (yellow box, driven by TK promoter) outside of 3′ homology arm is designed for negative selection. The sgRNA sequence (underlined in blue) is designed so that it spans across the stop codon (TAG, in bold) of the endogenous NEUROG2. Protospacer adjacent motif (PAM) sequence is underlined in red. Pink line in the schematic represents the homology arms. Restriction enzyme EcoRI is used to digest the genomic DNA of hiPSC reporter clones for Southern blot analysis. AscI is used in the intermediate steps of creating the targeting (donor) vector. ATG designates the start codon of NEUROG2 gene. Both 5′ and 3′’ flanking probes are designed for Southern blot analysis to identify correctly targeted hiPSC clones. The correctly targeted knockin (KI) allele is 8.4 kb and the untargeted allele [wild-type (WT)] is 11.6 kb. (B) SURVEYOR assay of sgRNA-mediated cleavage in 293FT cells by the CRISPR/Cas9 system. SURVEYOR assay is used to determine whether a designed sgRNA will be able to mediate specific excision of Cas9 at desired location. In this case, the uncut band is 502 bp and the cut bands are ∼360 and 142 bp. The sgRNA illustrated in (A) results in an indel (insertion or deletion) rate of ∼7.4%. Green fluorescent protein (GFP) plasmid is used as a control. (C) Southern blot analysis for conventional (designated as HRN clones) and CRISPR/Cas9-mediated NEUROG2 targeting (designated as CAN clones). Genomic DNAs from selected clones are digested with EcoRI and hybridized with 5′ or 3′ flanking probes. Data from 5′ probe hybridization for selected clones are shown here in (C). Data from 3′ probe hybridization for selected clones are shown in Supplementary Fig. S1. For the 35 clones that are selected from the conventional straight gene targeting, only one clone (HRN2), is correctly targeted (targeting efficiency 1/35, 2.9%). For the 42 clones that are selected from the CRISPR/Cas9-mediated gene targeting, 14 clones are targeted (targeting efficiency 14/42, 33.3%). * And ** designate the targeting at one and two alleles respectively. Δ, altered band size at one allele. (D) Sequencing results of targeted clones at the junction of NEUROG2 and IRES. CCA is the PAM sequence, TAG is the stop codon. Clones obtained through conventional gene targeting (HRN2) and CRISPR/Cas9-mediated gene targeting (CAN60, 82, and 127) show identical sequence. Partial sequence of AscI and SwaI sites are introduced during the cloning of IRES to donor vectors. Color images available online at www.liebertpub.com/scd