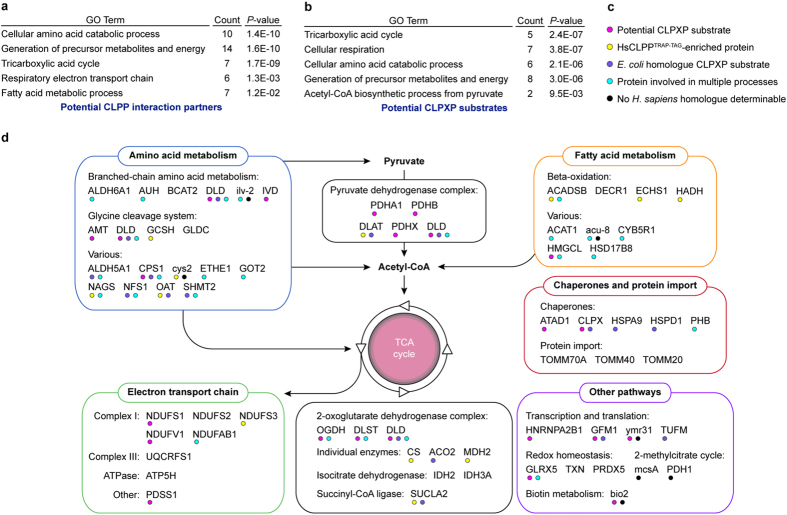
Figure 3. Potential mitochondrial CLPP interaction partners and CLPXP substrates.
(a) Selected GO terms enriched among human homologues of genes coding for proteins co-purifying with both HsCLPP variants (HsCLPPWT-/TRAP-TAG-shared) that were classified as potential CLPP interaction partneres. Count refers to the number of genes associated with the respective GO term. Only GO terms with Bonferroni-corrected P-values <5.0E-02 were considered statistically significant. For full GO enrichment analysis data see Supplementary Table 4. (b) Selected GO terms enriched among human homologues of genes coding for potential CLPXP substrates. For full GO term enrichment analysis data see Supplementary Table 5. c, Legend for colour-coded circles indicating different characteristics of the identified proteins. (d) Overview of proteins identified as HsCLPPWT-/TRAP-TAG-shared (Table 1), HsCLPPTRAP-TAG-enriched (Table 1) or potential CLPXP substrates (Table 2). Proteins are denoted by their corresponding gene names which are listed in Tables 1 and 2. Colour-coded circles below each protein indicate its respective characteristics. For example, the TCA cycle enzyme dihydrolipoyl dehydrogenase (DLD; Swiss-Prot ID: P09622; Table 2) was identified as a potential CLPXP substrate in our analysis (magenta circle), its bacterial homologue is a known E. coli CLPXP substrate (purple circle), and it is involved in multiple cellular processes (blue circle). Proteins with no associated circles, e.g. the mitochondrial peroxiredoxin-5 (PRDX5; Swiss-Prot ID: P30044; Table 1), were identified as potential CLPP interaction partners (i.e. they were specifically co-purified with both HsCLPP variants) but do not possess any of the additional characteristics.