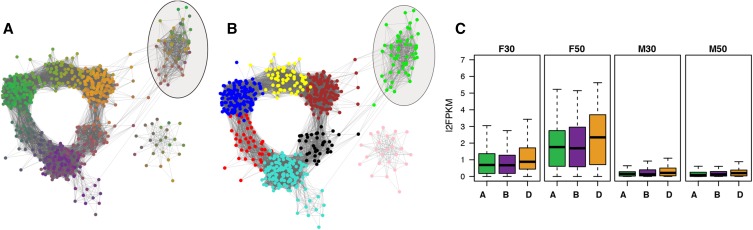
Figure 6.
Coexpression analysis of homeologous triplet genes. A set of conserved triplet genes, with one copy per subgenome (A, B, and D) was used to investigate genome-specific expression behavior and dosage effects in a triplet-based coexpression network. (A) Coloring the network nodes by expression contributions from individual genomes highlighted regions where the combined triplet expression is dominated by a single or two genomes. (B) The triplet network was split into triplet network modules with specific expression patterns for genome expression bias (Figure S11). (C) The boxplots show subgenome-wise expression strength of triplet members in a F. graminearum-responsive triplet module (highlighted in A and B) under the given conditions for NIL1 (Fg: treatment with F. graminearum; M: mock treatment; 30: 30hpi; 50: 50 hpi). Within this module, expression in response to the fungus was dominated by the D subgenome, which was also observed for NIL2, NIL3, and NIL4 (Figure S12).