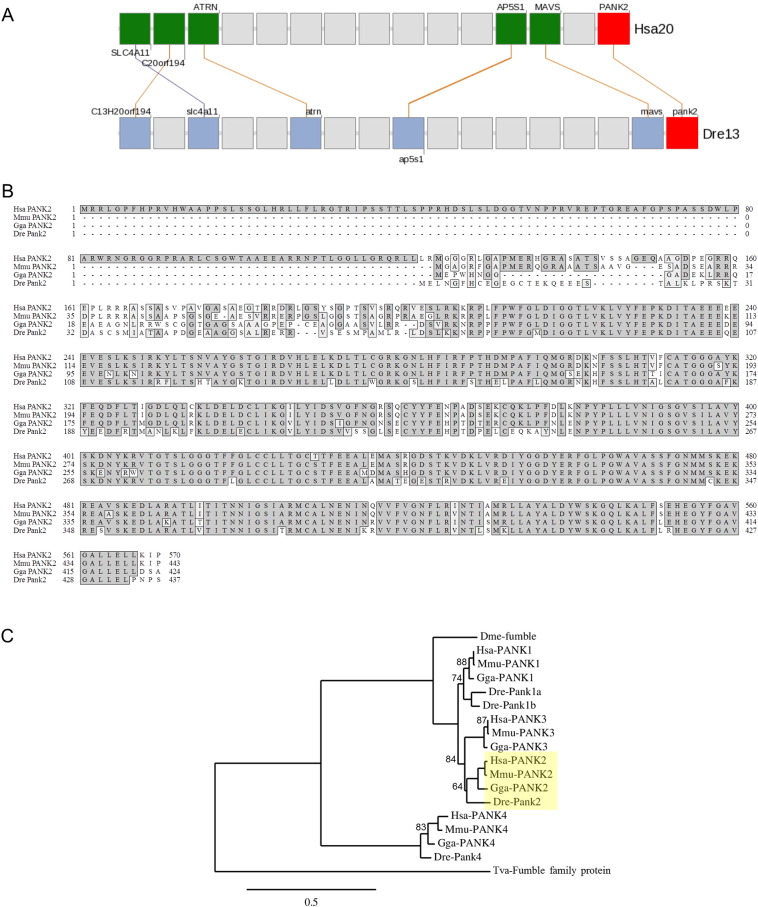
Fig. S1.
Bioinformatic analysis of zebrafish pank2 gene and protein sequences. (A) Gene trace synteny analysis around the PANK2 locus between Homo sapiens chromosome 20 (Hsa20) and Danio rerio chromosomes 13 (Dre13) generated using the Synteny Database (50-gene sliding window). Genes are drawn as squares, with their order preserved. Colored squares are members of the cluster while gray squares represent genes in the interval but that do not have orthologs or paralogs in the other segment. PANK2 genes are drawn as red squares. Lines connecting squares between the two clusters represent putative orthologous gene pairs.
(B) Multiple sequences alignment of human (Hsa) mouse (Mmu), chicken (Gga) and zebrafish (Dre) PANK2 proteins obtained using the ClustalW2 software. Residues that are identical are shown on a dark gray background; those on a light gray background are the conservative substitutions.
(C) Rooted tree showing phylogenetic analysis results for human (Hsa), mouse (Mmu), chicken (Gga) and zebrafish (Dre) pantothenate kinase family of proteins, as well as the fumble polypeptides from Drosophila melanogaster (Dme) and Tricomonas vaginalis (Tva). The tree clade including PANK2 polypeptides is highlighted in yellow. The horizontal bar represents a distance of 0.5 substitutions per site. Branch support values are displayed in percentage (support values > 90% are not shown). The tree, generated using the Phylogeny.fr web service, was rooted using the Tricomonas vaginalis Fumble family protein as an outgroup.