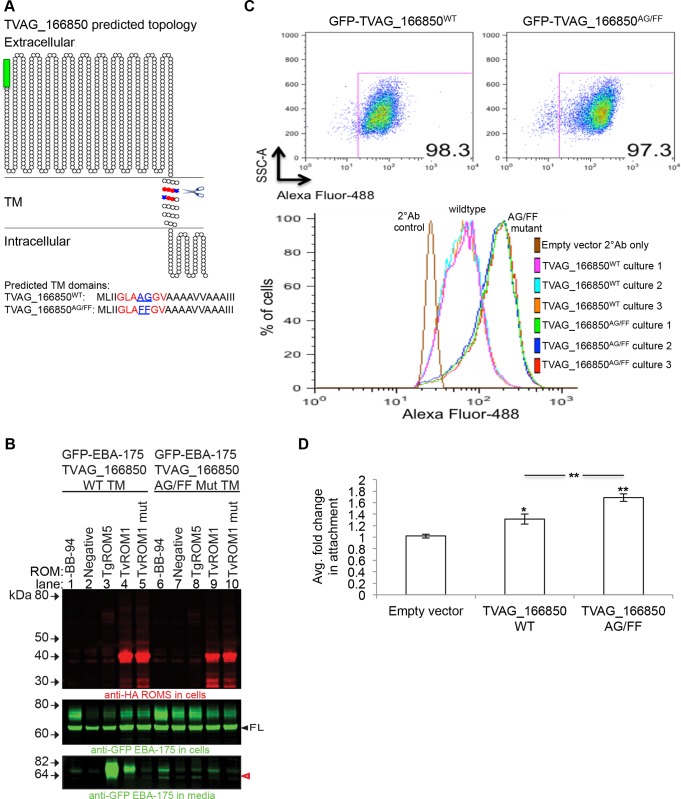
Fig 6. Phenotypic analysis of predicted rhomboid cleavage site mutation in the putative substrate TVAG_166850.
(A) Predicted topology of the putative substrate TVAG_166850 using the Spoctopus TM prediction program, illustrated using the TOPO2 graphical representation program. TVAG_166850WT was tagged at the N-terminus with a GFP tag (green box). The predicted rhomboid cleavage site (scissors) and the predicted P1-P1’ cleavage site residues are highlighted in blue. The surrounding parasite search motif residues are highlighted in red. The predicted TM residues are shown below. A rhomboid cleavage site mutant was generated by mutating the predicted Ala-Gly P1-P1’ residues (underlined in sequence) to Phe-Phe residues to generate the GFP-TVAG_166850AG/FF mutant. (B) TvROM1 efficiently cleaves the wild type (wt) TVAG_166850 TM and not the TVAG_166850AG/FF mutant TM. Proteases and chimeras of GFP-EBA-175 with the TVAG_166850 wt TM or mut TM were co-transfected in the heterologous cell cleavage assay. Lanes 1–5 = co-transfection with wild type GFP-EBA-175-TVAG_166850WT TM; lanes 6–10 = co-transfection with GFP-EBA-175-TVAG_166850AG/FF mutant TM. Western blot analyses of whole cell lysates confirm expression of wt and mut rhomboid proteases (top panel) and the full length substrates (FL, middle panel). Analyses of conditioned media show a GFP-tagged cleavage product in the media (red, open arrowhead, bottom panel) of TvROM1 co-transfected with wild type TVAG_166850 (lane 4) which is reduced ~90% in mutant TVAG_166850AG/FF co-transfectants (lane 9). Co-transfection with mutant TvROM1 abolishes cleavage (lanes 5 & 10). Lanes 1 & 6 = no metalloprotease inhibitor BB-94, lane 2–5 & 7–10 = 10 μM metalloprotease inhibitor BB-94. Lanes 3 & 8 = co-transfection with TgROM5 (control). (C) The GFP-TVAG_166850WT and GFP-TVAG_166850AG/FF mutant proteins were exogenously expressed in T. vaginalis. Transfectants were stained with an anti-GFP antibody without permeabilization at 4°C to detect surface levels of the fusion protein quantified using flow cytometry. Three cultures (cultures #1–3) in three independent experiments were analyzed. Representative results from one experiment are shown. Top panel shows the GFP+ cell population, similar percentages of GFP+ cells were detected for the wt and mutant transfectants. Bottom histogram shows the fluorescence intensity distribution of the GFP+ population. GFP-TVAG_166850AG/FF mutant transfectants had at least three-fold higher mean fluorescence intensity (MFI) levels compared to wild type transfectants. (D) The ability of GFP-TVAG_166850WT and GFP-TVAG_166850AG/FF mutant transfectants to attach to ectocervical cells was compared to empty vector transfectants. Results show the average of three experiments, each conducted in triplicate. Exogenous expression of GFP-TVAG_166850WT leads to a statistically significant increase in attachment compared to empty vector transfectants (*p<0.05). Overexpression of the GFP-TVAG_166850AG/FF mutant leads to an even greater increase in attachment compared to empty vector (**p<0.01) and wildtype GFP-TVAG_166850WT transfectants (**p<0.01).