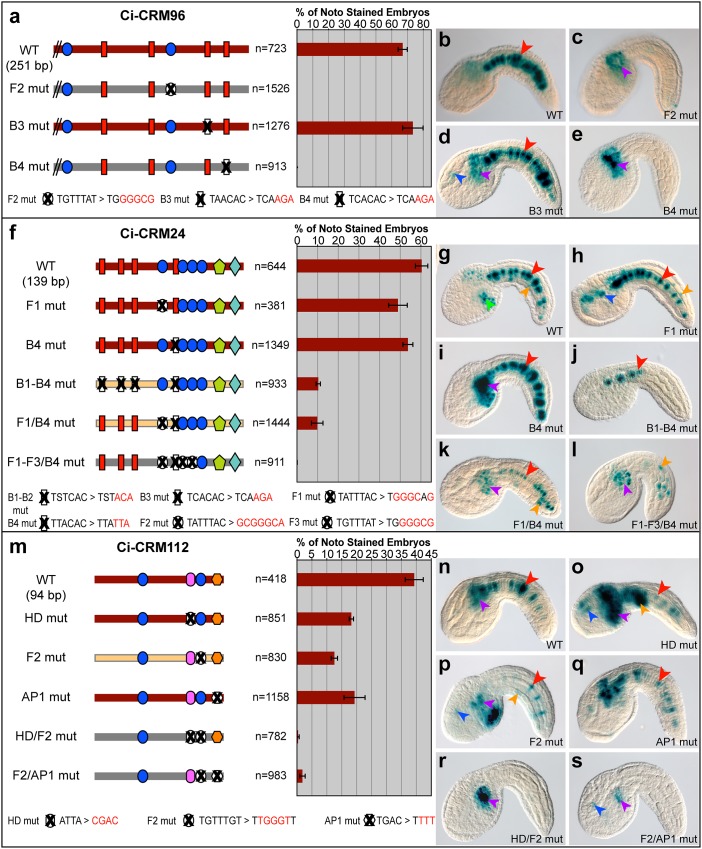
Fig 2. Alternative regulatory mechanisms of notochord CRMs requiring Ci-Bra and/or Ci-Fox binding sites.
a,f,m: (Left) Schematic representations of wild-type (WT) and site-directed mutant CRMs; TF binding sites are as in Fig 1, with the mutant sequences indicated at the bottom of each panel. Mutated binding sites are colored in white and covered by “X” signs. Maroon bars represent constructs able to elicit notochord expression, while configurations exhibiting weak or no notochord staining are depicted by yellow and gray bars, respectively. (Right) Quantification of the fraction of the total stained embryos showing notochord expression after electroporation of the constructs at the left of each bar. n: number of fully developed stained embryos. Error bars denote standard deviation from the mean. b-e, g-l, n-s: Microphotographs of embryos expressing the transgenes indicated at the bottom right of each panel. Arrowheads are color-coded as in Fig 1. Abbreviations: WT: wild-type, F: Fox binding site, B: Brachyury binding site, HD: homeodomain, AP1: activator protein 1, mut: mutated, noto: notochord. In f, “S” stands for C/G. See also S2 Fig.