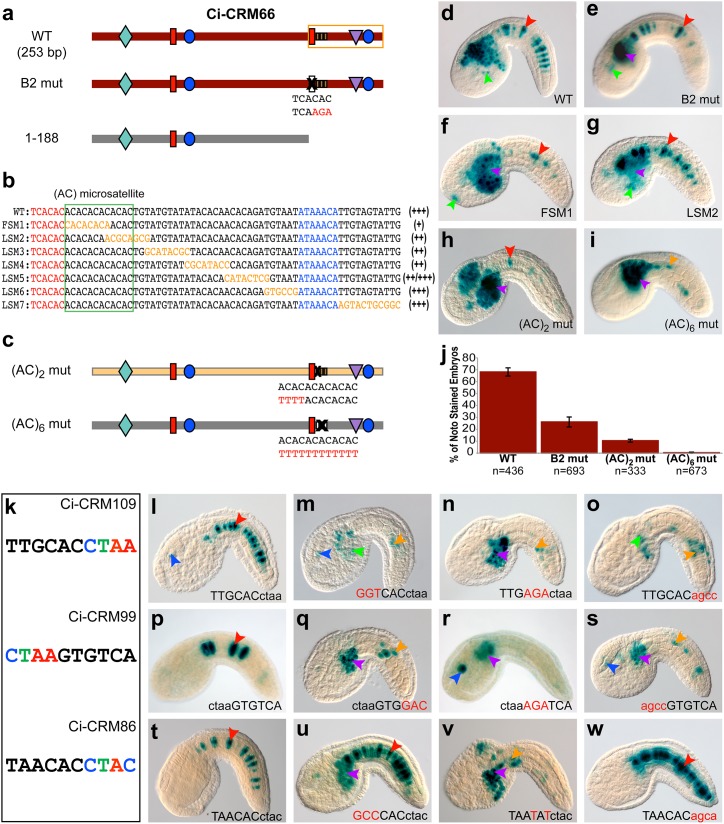
Fig 3. The function of individual Ci-Bra binding sites can be modulated by either an (AC) microsatellite or a flanking sequence.
a,c: Schematic representations of wild-type (WT) and mutant CRMs, as described and colored in Fig 2; the (AC) microsatellite sequence is schematized as a segmented brown rectangle. b: Mutational series of the area boxed in orange in the 253-bp construct. Red and blue nucleotides correspond to the Ci-Bra and Ci-Fox sites, respectively, and orange nucleotides indicate the bases changed in each mutant plasmid. The (AC)6 microsatellite sequence is boxed in green. The relative ability of each construct to direct notochord gene expression is shown by plus signs at the right of each sequence. d-i: Photos of embryos electroporated with the constructs depicted in a,b,c; arrowheads are color-coded as in Fig 1. j: Quantification of notochord-stained embryos harboring the constructs in a,c. Error bars indicate standard deviation from the mean. k: Identification of an extended CTAM sequence (colored) shared by a subset of individually-acting Ci-Bra binding sites. l-w: Microphotographs of embryos carrying wild-type CRMs (l,p,t) compared to embryos carrying various mutant versions of Ci-CRM109 (m-o) Ci-CRM99 (q-s) and Ci-CRM86 (u-w). Core Ci-Bra binding sites are capitalized. Mutations are depicted in red. Abbreviations: FSM: “frame-shift” mutation, LSM: linker scanning mutation. See also S3 Fig.