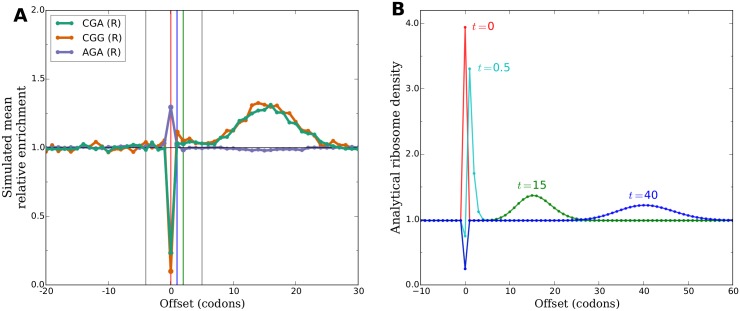
Fig 4. A change in the relative elongation rates of codons produces downstream waves in simulation and analytical models.
(A) In a simulation of the translation of yeast coding sequences, the average relative elongation time of each codon identity was changed from the codon’s A-site enrichment in a no-CHX experiment to its A-site enrichment in a CHX experiment. Elongation was allowed to proceed for a short time after this change, then enrichments in the positions of ribosomes were analyzed as in Fig 3B. The resulting profiles of simulated mean enrichments qualitatively reproduce the downstream peaks in data from experiments using CHX. (B) A model of the translation of a hypothetical coding sequence consisting of a single slow codon surrounded by long stretches of identically faster codons on either side was analyzed. At t = 0 (arbitrary units), the relative speed of the slow codon was changed to be faster than its surroundings. Ribosome density at offsets around the formerly-slow codon is plotted at several time points after this change. Immediately after the change, there is a temporary excess of ribosomes positioned at the formerly-slow codon relative to the eventual steady state of the new dynamics. As these excess ribosomes advance along the coding sequence, a transient wave of increased ribosome density moves downstream and spreads out over time.