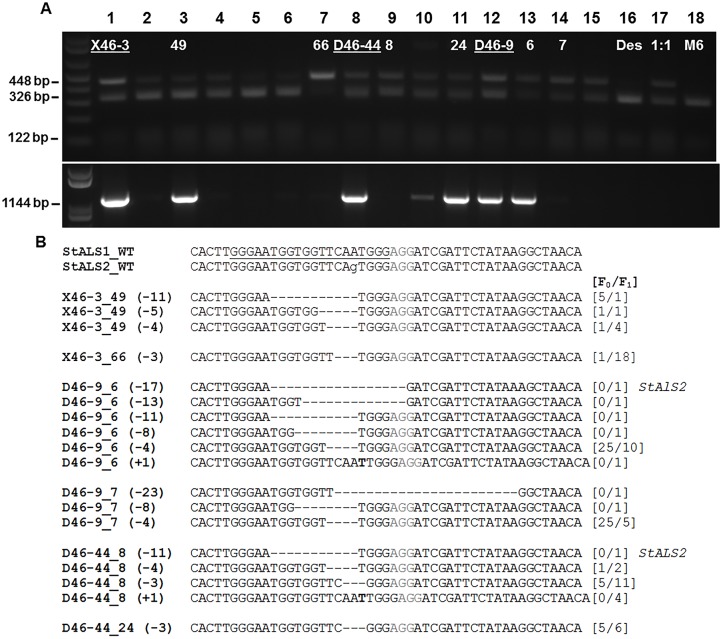
Fig 3. Inheritance of targeted mutations and Cas9 in progeny of primary CRISPR/Cas events.
Three primary events with cloned targeted mutations (lanes 1, 8 and 12; underlined) were used to generate genetic populations to assess inheritance of targeted mutations. The diploid event (lane 1; X46-3) was crossed to an inbred diploid line, M6 (lane 18) as the female parent while tetraploid events (lanes 8, 12; D46-44, D46-9) were selfed. Six progeny from the X46-3 population (lanes 2–7) and three progeny from the D46-44 (lanes 9–11) and D46-9 (lanes 13–15) populations were assessed for A) targeted mutations using a restriction digestion assay (top gel) and inheritance of Cas9 (bottom gel) and used for B) cloning targeted mutations using previously described methods (Fig 2 and S5 Fig). The PCR assay used for detecting Cas9 (A; bottom gel) produced a 1144 bp amplicon with each lane corresponding to the top gel and is further described in S6 Fig Wild-type Désirée and M6 were used as negative controls (lanes 16 and 18, respectively) and a 1:1 template mixture with wild-type and mutated DNA was used as a positive control (lane 17). The lengths of deletions (-) or insertions (+) of the targeted mutations in progeny (B) are in parenthesis to the left of each cloned mutation and the number of reads generated in the primary event (F0) or individual progeny (F1) are in brackets on the right. All targeted mutations were aligned to wild-type sequence and cloned from StALS1 unless indicated on the right. PAM sequences are in gray.