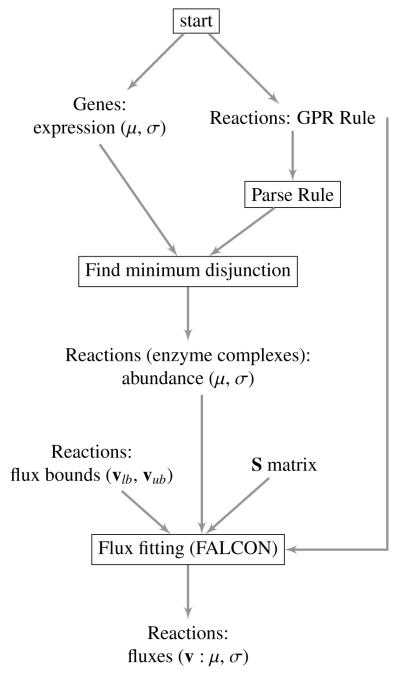
Figure 1.
Flowchart illustrating the two algorithms used in this paper. The process of estimating enzyme complex abundance is displayed in detail, whereas the flux-fitting algorithm (FALCON) is illustrated as a single step for simplicity. First, for each gene in the model with available expression data, the mean and (if available) standard deviation or some other measure of uncertainty are read in. Gene rules (also called GPR rules) are also read in for each enzymatic reaction. The reaction rules are parsed and the minimum disjunction algorithm is applied, making use of the gene’s mean expression. Next, the estimated and unitless enzyme complex abundance and variance are output for each enzymatic reaction. Finally, flux fitting with FALCON (Algorithm 3.1) can be applied, and requires the model’s stoichiometry and flux bounds. The final output has the option of being a deterministically estimated flux, or a mean and standard deviation of fluxes if alternative optima are explored.