Abstract
The Liver X Receptors (LXRs) play important roles in multiple metabolic pathways, including fatty acid, cholesterol, carbohydrate and energy metabolism. To expand the knowledge of the functions of LXR signaling during embryonic development, we performed a whole-genome microarray analysis of Lxr target genes in zebrafish larvae treated with either one of the synthetic LXR ligands T0901317 or GW3965. Assessment of the biological processes enriched by differentially expressed genes revealed a prime role for Lxr in regulating lipid metabolic processes, similarly to the function of LXR in mammals. In addition, exposure to the Lxr ligands induced changes in expression of genes in the neural retina and lens of the zebrafish eye, including the photoreceptor guanylate cyclase activators and lens gamma crystallins, suggesting a potential novel role for Lxr in modulating the transcription of genes associated with visual function in zebrafish. The regulation of expression of metabolic genes was phenotypically reflected in an increased absorption of yolk in the zebrafish larvae, and changes in the expression of genes involved in visual perception were associated with morphological alterations in the retina and lens of the developing zebrafish eye. The regulation of expression of both lipid metabolic and eye specific genes was sustained in 1 month old fish. The transcriptional networks demonstrated several conserved effects of LXR activation between zebrafish and mammals, and also identified potential novel functions of Lxr, supporting zebrafish as a promising model for investigating the role of Lxr during development.
Keywords: Zebrafish, Liver X receptor, Gene Regulation, Lipid Metabolism, Cholesterol Metabolism, Eye, Retina, Lens, Yolk
1. Introduction
Liver X receptors (LXRs) belong to the nuclear receptor subfamily of ligand-activated transcription factors. Two LXR genes are present in mammals, LXRα (NR1H3) and LXRβ (NR1H2), encoding highly related proteins that share ~78% amino acid sequence identity in both DNA and ligand-binding domains. Both LXR proteins form permissive heterodimers with retinoid X receptor (RXR) and control gene expression through binding to LXR response elements (LXREs) in the promoter region of target genes (1, 2). LXRs are key regulators of multiple metabolic pathways, including fatty acid, cholesterol, carbohydrate and energy metabolism, and they also have regulatory roles in inflammatory responses and immunity (3). The endogenous LXR agonists are oxidized derivatives of cholesterol referred to as oxysterols. Synthetic ligands, such as T0901317 (T0) and GW3965 (GW), can also modulate LXR transcriptional activity (3-5); however, T0 is also known to be a ligand for other nuclear receptors, such as farnesoid X receptor (FXR) and pregnane X receptor (PXR) in humans (6).
During embryonic development in rodents, LXRα is expressed in the liver, yolk sac, small intestine and other tissues involved in lipid metabolism. LXRβ is also detected in the embryonic liver and endocrine tissues, but is additionally expressed early on in several structures of the CNS, including the retina (7, 8). In adulthood, LXRα transcript remains high in tissues involved in metabolism, whereas LXRβ is ubiquitously expressed (7).
In addition to their role in regulation of lipid metabolism in mammals, LXRs also have functions in the CNS (9). In vivo studies have shown that LXRs promote ventral midbrain neurogenesis and dopaminergic neuron development (10), and that they are implicated in the differentiation process of Bergmann glia cells, and migration of neurons during cerebellar development (11). LXRβ is also important for cerebral cortex lamination and neocortical neuron migration in mice (12). However, a potential function for LXR in the retina and other neuronal structures remains obscure.
The role of Lxr has also been studied in zebrafish. While there are two LXR genes in mammals, only one lxr gene with higher similarity to the mammalian LXRα is present in zebrafish. During zebrafish development, lxr transcripts are detected in several tissues, such as the liver, intestine, brain, neural retina and lens, and its expression pattern remains ubiquitous at the adult stage (13, 14). Similarly to its role in mammalian systems, lxr also seems to play a crucial role in the control of lipid homeostasis in zebrafish. Previous studies have indicated that Lxr activation with synthetic ligands induced the expression of genes related to cholesterol transport and lipid synthesis in the liver, brain and eye of adult zebrafish (13-15). Additionally, treatment with the Lxr agonist T0 induced hepatic lipid accumulation in adult zebrafish, an effect that has also been shown in mammalian models (15, 16).
Studies on the role of Lxr in zebrafish during embryonic development are scarce (13, 14). The expression of genes important for cholesterol transport and lipid synthesis has been shown to be affected in zebrafish embryos exposed to GW, and morpholino knockdown of Lxr resulted in impaired lipid deposition in the head and eye region of the zebrafish embryos, malformation of the pharyngeal skeleton and in elevated levels of cholesterol in the yolk of the Lxr morphant (13), suggesting an important function of Lxr in controlling lipid homeostasis in the developing zebrafish. However, more studies are needed to assess the molecular networks affected by Lxr activation in zebrafish at early developmental stages.
In this report, we investigated whether Lxr has a primary function in regulating lipid metabolic processes during zebrafish development, similarly to the role of LXR in mammals. We also set out to discover new functions of Lxr during zebrafish development. We first characterized the ability of the synthetic human LXR ligands T0 and GW to activate zebrafish Lxr by generating a stable reporter cell line expressing the zebrafish Lxr ligand binding domain. Next, we performed a whole-genome microarray analysis on zebrafish larvae treated with the synthetic ligands to evaluate organism wide transcriptional effects induced by Lxr activation during zebrafish larval development. Assessment of enriched biological processes provided insights into the molecular pathways altered by Lxr activation during zebrafish development, and tissue enrichment analysis inferred the tissues most affected by Lxr-mediated signaling in zebrafish. The expressions of genes not previously shown to be altered by Lxr activation in vertebrates were revealed in zebrafish. This study supports zebrafish as an alternative in vivo model for Lxr-related studies and demonstrates a novel role for Lxr during development.
2. Material and Methods
2.1. Cell culture conditions and generation of stably transfected reporter cell lines expressing GAL4-zfLxr and GAL4-zfPparγ
The cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with phenol red and supplemented with 5% fetal calf serum (FCS), 1% glutamine, 1% penicillin/streptomycin and the selection agents puromycin (0.5 μg/mL) and G418 (1 mg/mL) in a 5% CO2 humidifed atmosphere at 37°C. All cell culture reagents were obtained from Gibco (Grand Island, NY).
The generation of the reporter cell line expressing the zebrafish Lxr (HG5LN-zfLxr cells) was performed in two steps. First, HG5LN cell line was generated by transfecting HeLa cells with p(GAL4RE)5-βGlob-Luc-SVNeo plasmid as previously described (17). Second, the HG5LN cells were stably transfected with pGAL4-zfLxr-puro plasmid. To construct GAL4-zfLxr chimeras, the LBD of zebrafish Lxr (aa 175 to aa 412) was amplified by PCR with primers containing XhoI and BamHI restriction sites, and the PCR fragment was used to substitute the glucocorticoid receptor (GR) LBD in the pGAL4-GR-puro plasmid as described by Seimandi et al, 2005. Cells were transfected by electroporation and selected with 0.5 μg/ml puromycin (Sigma-Aldrich). Three weeks after initiation of puromycin selection, each individual clone was transferred to one well in 96-well tissue culture plates (Greiner Bio-One, NC, USA). Luminescence emitted from the individual clones prior to and after overnight treatment with 1 μM T0 (T0901317, Sigma-Aldrich, St. Louis, MO) was measured with medium containing 0.3 mM luciferin (Promega, Charbonnieres les Bains, France). The clones with best induction factor were selected and expanded in culture in the presence of 0.5 μg/mL puromycin. Generation of luciferase reporter cells expressing zebrafish Pparγ (HG5LN-zfPparγ cells) has been described previously (18).
2.2. Luciferase assays
HG5LN-zfLxr and HG5LN-zfPparγ cells were seeded at 40,000 cells per well in 96-well white opaque tissue culture plates, and maintained in DMEM supplemented with 5% FCS. Twenty-four hours later, culture medium was replaced with phenol-red and serum-free DMEM containing T0 or GW (GW3965, Tocris, Bristol, UK), and cells were incubated with the ligands for 16 hours. At the end of the incubation, culture medium was replaced with fresh medium containing 0.3 mM luciferin, and luciferase activity was measured for 2 seconds using the Victor X4 Multilabel Plate Reader (Perkin Elmer, Waltham, MA). Nonlinear regression curve fit was used to plot the dose response curves using GraphPad Prism version 5 (GraphPad Software, La Jolla, CA). Experiments were performed in quadruplicate and repeated three times.
2.3. Zebrafish maintenance
Wild type WIK and AB/Tuebingen (TAB) were housed at 28.5°C in a Tecniplast system (Tecniplast USA Inc., West Chester, PA) on a 14 hour/10 hour light/dark cycle. They were used according to the maintenance and experimental protocols approved by the Institutional Animal Care and Use Committee at University of Houston (protocol numbers 12-042 and 13-028).
2.4. Zebrafish larvae treatments for microarray analysis
Wild type zebrafish WIK embryos were collected after spawning. At 4 days post fertilization (dpf), zebrafish larvae were transferred to 6-well plates (pools of 30 embryos/well) in 3 mL of embryo media (E3: 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.3 mM MgSO4). Larvae at 4 dpf were treated with DMSO (Sigma-Aldrich, St. Louis, MO) as vehicle control, 2 μM T0 or 1 μM GW. Final DMSO concentration in all treatments and control was 0.1%. Treatments were renewed after 24 hours and larvae were collected for RNA extraction at 6 dpf. Treatments with vehicle (0.1% DMSO), T0 and GW were performed in biological quadruplicates of pools of RNA. Confirmation of selected genes by RT-qPCR was performed in 3-4 independent pools of larvae exposed to the LXR ligands.
2.5. Juvenile Zebrafish treatments
Wild type (TAB14) zebrafish at the juvenile stage (1 to 3 month-old) were kept in Falcon tubes (Corning Life Sciences, Tewksbury, MA) at the density of 5 fish per tube in 25 mL water, and treated daily for 4 days with either DMSO 0.1% as vehicle control or with 1 μM GW or 1 µM TO. Fish were fed once a day with baby brine shrimp (Brine Shrimp Direct, Ogden, UT) for 30 minutes. At the end of the treatments, fish were anesthetized with 0.04% tricaine methylsulfonate (MS-222, Sigma-Aldrich) and RNA from single whole fish was extracted for cDNA synthesis.
2.6. RNA extraction and cDNA synthesis
Total RNA from pooled larvae for the microarray experiments, and RNA from whole single juvenile fish were extracted using Trizol (Invitrogen Corporation, Carlsbad, CA) and RNeasy spin columns (Qiagen, Chatsworth, CA) according to the manufacturer’s protocol. On-column DNase I (Qiagen, Chatsworth, CA) digestion was performed to remove remaining DNA. RNA concentrations were measured with NanoDrop 1000 spectrophotometer (Agilent Technologies, Palo Alto, CA) and Superscript III reverse transcriptase (Invitrogen Corporation, Carlsbad, CA) with random hexamer primers were used for cDNA synthesis.
2.7. Microarray scanning and analysis of resulting data
Agilent zebrafish (V3) gene expression microarray 4X44K (Design ID 026437) was used for the microarray analysis. Labeling of samples, followed by hybridization and scanning of arrays was performed by the Genomic and RNA Profiling Core at Baylor College of Medicine (Houston, TX) according to protocol previously described (19). Microarray data was processed in the R environment for statistical computing using the Limma package for analysis of differential expression (Smyth 2005). Arrays were quantile normalized, replicated spots averaged and empirical Bayes methods applied for the differential-expression analysis. Four arrays of DMSO and GW-treated zebrafish and three arrays of T0-treated zebrafish were used for microarray data analyses. Statistical significance was assessed using false-discovery rate (FDR) 5%. The resulting dataset with FDR-adjusted P≤0.05 and absolute fold-change (FC) |±1.4| were subjected to hierarchical clustering using complete-linkage clustering of the Euclidean distance measure of the fold change values in each sample, and Venn diagrams were generated to represent the overlapping genes between the two treatments. Non-annotated probes were removed from the dataset, and probes mapped to the same gene were resolved using the minimun fold-change value to maintain a conservative analysis. The lists of statistically significant genes commonly and uniquely regulated by T0 and GW are displayed in the supplemental information, along with calculated fold-change and adjusted p-values (Tables S1, S2, S3). The microarray results have been submitted to the GEO database (Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo) (GEO accession # GSE59031).
2.8. Biological function and tissue enrichment analysis
Differentially expressed zebrafish genes (P≤0.05, fold change ≥ |±1.4|) were mapped to their respective human homologues using Homologene (www.ncbi.nlm.nih.gov/homologene), ZFIN (http://zfin.org/) and Ensembl (http://www.ensembl.org/). Pathway Studio software (Elsevier Inc. MD, USA) was used for the analyses of enriched biological processes for the human homologues of the zebrafish genes regulated by T0 and GW using Gene Ontology (GO) functional annotations. Enriched subnetwork analysis was applied to identify transcriptional regulators that are responsible for the gene expression profile obtained from microarray data. Fisher’s Exact test was applied to determine enriched GO functional groups and subnetworks; P≤0.05 was considered significant. REVIGO (http://revigo.irb.hr/) was used to summarize the list of enriched GO terms obtained after functional analysis of the whole set of differentially expressed genes by each ligand. GO terms with P<0.0001 and SimRel semantic medium (0.7) similarity were used for REVIGO clustering analysis.
In order to estimate the tissues mostly affected by LXR activation in zebrafish, tissue enrichment analysis was performed with zebrafish genes regulated by the LXR ligands using the ZFIN_Anatomy functional category at the NIH DAVID Bioinformatic Resources database (http://david.abcc.ncifcrf.gov/). Fisher’s Exact test was used to calculate enriched functional categories; enriched tissues with P≤0.05 and with a minimum of 5 enriched genes were considered.
2.9. Quantitative real-time PCR (RT-qPCR)
Confirmation of selected gene expression changes by qPCR were performed using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA) with iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) according to the manufacturer’s protocol. Primers for confirmation of selected genes were designed using Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and synthesized by Integrated DNA Technologies, Inc (Coralville, IA). The primer sequence list is provided in Supplemental Table 4. Transcript abundance was normalized to β-actin (bac2) as the endogenous reference gene and normalization of the data was done using the ΔΔCt method.
2.10. Oil Red O (ORO) Staining
Larvae were treated with the LXR ligands following the protocol used for the microarray experiment. At 6 dpf, larvae were euthanized with tricaine, fixed in 4% paraformaldehyde (PFA) overnight at 4°C, washed in PBT (PBS with 0.1% Tween 20) and stained with a filtered 0.3% Oil Red O (ORO) solution for 3 hours at room temperature. After staining, larvae were washed 3 times in PBT and stored in 90% glycerol before imaging. Larvae were scored for ORO staining in the yolk and imaged using a Nikon AZ100M microscope equipped with a color camera DS Fil (Nikon) and the NIS-Elements AR imaging software (Nikon Instruments Inc, Melville, NY).
2.11. Zebrafish eye histology
For the zebrafish eye histological studies, zebrafish embryos were treated daily starting at 6 hpf with either DMSO 0.1%, 2 μM T0 or 1 μM GW until 3 dpf, and at 4 dpf zebrafish embryos were euthanized, fixed in 4% PFA, washed in PBT, dehydrated in methanol and embedded in plastic using the JB-4 Plus Embedding Kit (Polysciences, Warrington, PA). Sections were cut at 8-15 μm and counterstained with hematoxylin and eosin (H&E staining). Sections were imaged using an Olympus BX51 microscope with the cellSens software (Olympus, PA).
2.12. Morpholino knock-down
Lxr knock-down was performed by microinjection of antisense morpholino oligonucleotides (MO) against lxr mRNA, which were designed and synthesized by Gene Tools, LLC (Philomath, OR). The sequences of the translational blocking lxr MO was 5’-TCTCCTGTTTCACTTCTGCCATCGC-3’ (−3 to 22, using the first nucleotide of the start codon as the reference). The sequence of the mismatched control MO for lxr containing 5 base substitutions (in lower case) was 5’-TaTCCTaTTTCAaTTCTGCaATCaC-3’ (−3 to 22). Microinjections were carried out using a microinjector (Harvard Apparatus, MA) into 1-2 cell stage wild-type zebrafish embryos. The dosage for MO injection was 1 ng per embryo. Injected embryos were processed at 4 dpf for RNA extraction and/or fixed for histology, as described above.
2.13. Analysis of Cell Death
For visualization of cell death in the eye of live zebrafish, zebrafish embryos were treated daily with LXR ligands starting at 6 hpf until 4 dpf, followed by staining with 10 mg/ml Acridine Orange (AO) for 30 minutes. Next, embryos were imaged on an Olympus IX51 inverted fluorescence microscope with the cellSens software (Olympus, PA), and the number of fluorescing spots in the eye region was recorded for each fish. The data was analyzed and plotted using GraphPad Prism version 5. Statistical analysis was performed using the Student’s t-test.
For histological analysis of cell death in the eye, zebrafish embryos were treated daily with LXR ligands from 6 hpf to 6 dpf . Embryos were euthanized after treatment, fixed in 4% PFA overnight, washed in PBS, dehydrated consecutively in 25% and 35% sucrose solutions, and embedded for cryosectioning. Sections were cut at 10-15 µm, and fixed in 4% PFA, washed three times in PBS, and permeabilized with 0.1% Triton-X-100 (Sigma) for 8 minutes at room temperature. Finally, sections were incubated with enzyme and labeling solution from an In situ cell death detection kit (Roche, IN ). Sections were washed in PBS, mounted with UltraCruz Mounting medium containing DAPI (Santa Cruz Biotechnology, TX), and imaged using an Olympus BX51 fluorescence microscope.
3. Results
3.1. The mammalian LXR agonists T0901317 (T0) and GW3965 (GW) activate zebrafish LXR
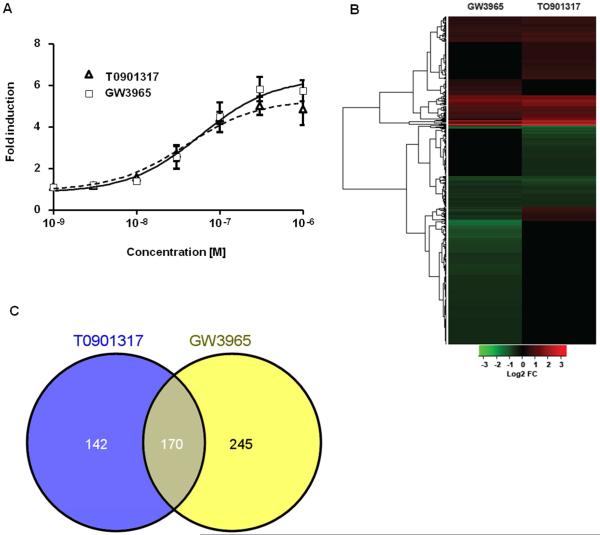
The LXR ligands T0 and GW are potent synthetic agonists of mammalian LXRs (16, 20). We constructed a stable zebrafish Lxr (zfLxr) reporter cell line, in which the lxr ligand binding domain was fused to the GAL4 DNA binding domain, and luciferase expression was driven by upstream UAS elements. Both T0 and GW activated zfLxr in a dose-dependent manner with EC50s of 39.15 nM for T0 and 58.01 nM for GW (Fig. 1A), in the same range of EC50 values published for the mammalian LXRs (16, 20). It is known that T0 is also a ligand for farnesoid X receptor (FXR) and pregnane X receptor (PXR) in humans, and for Fxr in zebrafish (6). Using our previously constructed zebrafish peroxisome proliferator gamma (zfPparγ) reporter cells (Riu et al. 2014), we found that GW, but not T0, activated zebrafish Pparγ-driven transcription (Supplemental Fig. S1). Considering that both T0 and GW did not specifically activate only Lxr, microarray analysis was performed in zebrafish larvae treated with either T0 or GW and the genes co-regulated by both ligands were used for subsequent biological functional analysis, as these genes most likely represent specific Lxr-mediated effects in zebrafish.
Figure 1. The LXR ligands T0901317 and GW3965 activate zfLxr and affect gene expression in zebrafish larvae.
Transactivation assays performed in HG5LN cells stably transfected with chimeric GAL4-zfLXR LBD in the presence of increasing concentrations of T0901317 and GW3965. Results are presented as mean ± s.d. of 3 independent experiments. (A) Heat map of differentially expressed genes in 6 dpf zebrafish larvae exposed to T0901317 or GW3965 as determined by microarray analysis (absolute fold change ≥ |±1.4|, FDR-adjusted P≤0.05) (B). Venn diagram illustrating the number of statistically significant genes that were co-regulated in zebrafish larvae treated with T0901317 or GW3965 (C).
3.2. Lxr target genes cluster in transport and metabolism of lipids
To assess the effects of Lxr activation during zebrafish development, zebrafish larvae were exposed to the Lxr agonists T0 (2 μM) or GW (1 μM) for two days starting at 4 dpf, with treatment renewal after 24 hours. The concentrations of the ligands were chosen based on a dose response activation of the known Lxr target genes abca1a, abcg1 and fasn in zebrafish larvae assessed by RT-qPCR (data not shown). At 6 dpf, RNA was extracted for microarray analysis. As represented in the hierarchical clustering of differentially expressed genes and in the Venn diagram (Figs. 1B,C), transcriptomic analysis revealed that the expression of a total of 170 genes was co-regulated by both Lxr ligands, whereas the expression of 142 and 245 genes were uniquely regulated by T0 and GW, respectively (P≤0.05, absolute fold change ≥ |±1.4|). Out of the 170 genes with mutually regulated expression by the ligands, the expression of 88 genes was upregulated, the expression of 63 genes was downregulated, and the expression of a set of 19 genes was affected in opposite directions (upregulated by T0 and downregulated by GW). The genes with altered expression by one or both of the ligands are shown in Supplemental Tables 1-3.
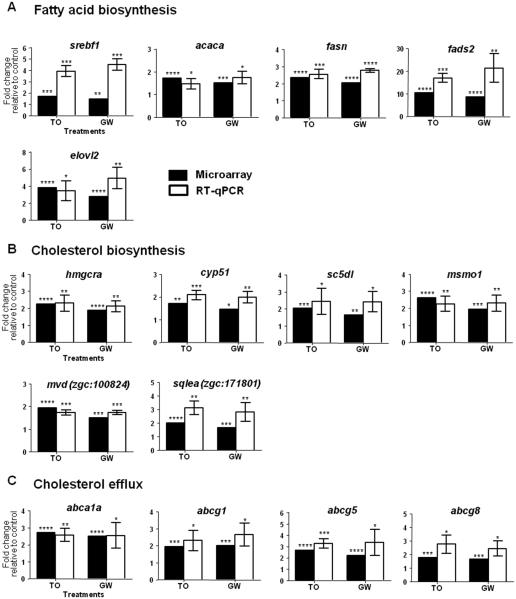
In order to predict the effects of Lxr-mediated gene regulation in zebrafish, the differentially expressed genes by T0 or GW were mapped to their human orthologues using ZFIN, Ensembl zebrafish Zv9, and NCBI HomoloGene databases, followed by Gene Ontology (GO) enrichment analysis of the human orthologues using Pathway Studio database. Table 1 displays the biological processes enriched by genes whose expression was co-regulated by both Lxr ligands. The most significantly enriched biological processes were all involved in the metabolism and biosynthesis of lipids, including cholesterol and fatty acid biosynthetic processes. Transcriptomic analysis showed upregulation of expression of genes related to de novo fatty acid synthesis (acaca, fasn, scd) and synthesis of long-chain polyunsaturated fatty acids (fads2 and elovl2) in zebrafish larvae treated with the Lxr ligands, further confirmed by RT-qPCR (Fig. 2A). Indeed, the most strongly induced gene by both T0 and GW was fads2, a fatty acyl desaturase, with absolute fold-changes of 10.4 and 8.5, respectively. Expression of srebf1 (sterol regulatory binding factor 1), coding for a transcription factor that activates genes involved in the synthesis of fatty acids and their incorporation into triglycerides and phospholipids, as well as genes involved in cholesterol biosynthesis, was also significantly induced in zebrafish exposed to the Lxr ligands (Fig. 2A). Moreover, microarray analysis revealed that the expression of several other gene products important for cholesterol biosynthesis was significantly induced by both Lxr ligands in zebrafish, and included srebf2 and key enzymes in the cholesterogenesis pathway (Table 1), out of which, six were selected for qPCR confirmation (Fig. 2B). The well-known mammalian LXR-regulated processes cholesterol homeostasis and efflux were also enriched in the functional analysis. The expression of ATP-binding cassette transporters abca1a, abcg1, abcg5 and abcg8, all classical LXR target genes and involved in cholesterol reverse transport, were induced in zebrafish larvae exposed to the ligands (Fig. 2C).
Table 1.
Top ranked biological processes enriched by genes co-regulated in zebrafish larvae exposed from 4 dpf to 6 dpf to 2 μM T0 or 1 μM GW.
| Enriched biological process |
Zf Gene Symbol | p-value | GO ID |
|---|---|---|---|
| Lipid metabolic process |
apoea,abca1a,abcg1, apoa4, ldlrb, abcg8, srebf1, abcg5, scd,
acaca, lipeb, dgat2, nr1h3, pltp, insig1, acsl3b, acsl4a, lipg, fads2, lipin2, ch25hl1.1, echs1, sc5dl, sult2st1, soat2, lipi, acsbg2 |
2.77E-27 | 6629 |
| Lipid biosynthetic process |
hmgcra, srebf1, scd, acaca, fasn, dgat2, elovl2, cyp51, mvd, lss, hsd17b7,
fads2, ch25hl1.1, msmo1, ebp, sc5dl |
2.79E-20 | 8610 |
| Cholesterol homeostasis |
apoea,abca1a,abcg1, ldlrb, apoa4, abcg8, abcg5, g6pca.1, nr1h3, lipg,
mylipa, soat2 |
5.51E-18 | 42632 |
|
Cholesterol metabolic
process |
apoea, abca1a, abcg1, ldlrb, apoa4, srebf1, lipeb, insig1, sqlea,
ch25hl1.1, ebp, soat2 |
2.10E-16 | 8203 |
|
Cholesterol biosynthetic
process |
hmgcra, insig1, cyp51, mvd, sqlea, lss, hsd17b7, msmo1, ebp, scd5l | 4.33E-16 | 6695 |
| Sterol biosynthetic process | hmgcra, insig1, cyp51, mvd, sqlea, ch25hl1.1, msmo1, ebp, scd5l | 1.28E-15 | 16126 |
| Response to nutrient |
bcl2, hmgcra, abca1a, abcg8, abcg5, aglb, acsl3b, acsl4a, mgp,
a2ml, lipg, slc6a19a, soat2 |
3.68E-15 | 7584 |
| Cholesterol efflux | apoea, abca1a, abcg1, apoa4, abcg8, abcg5, abcg4, soat2 | 5.79E-14 | 33344 |
|
Steroid biosynthetic
process |
hmgcra, prlra, cyp51, mvd, lss, hsd17b7, ch25hl1.1, msmo1, ebp, scd5l | 8.07E-13 | 6694 |
| Metabolic process |
abca1a, abcg1, prkg1a, atp1a1a.2, ca4c, aanat2, abcg5, scd, acaca,
g6pca.1, lipeb, fasn, insig1, ids, cox6a2, acsl3b, acsl4a, zgc:101000, hsd17b7, fads2, sqlea, mylipa, abcd3a, cdc42bpa, ch25hl1.1, msmo1, ebp, atp6v1e1b, echs1, pank1a, grhprb, dera, ggctb, acsbg2, agxt2l1 |
1.02E-12 | 8152 |
|
Fatty acid biosynthetic
process |
scd, acaca, fasn, nr1h3,elovl2, acsl3b, fads2,ch25hl1.1, msmo1,scd5l | 1.30E-12 | 37 |
Bold font indicates genes significantly upregulated by both LXR ligands, while downregulated genes are in regular font. Underlined genes were upregulated by T0 and downregulated by GW.
Figure 2. RT-qPCR validation of selected genes and comparison with microarray data.
qPCR analysis was performed on selected genes involved in fatty acid biosynthesis (A), cholesterol biosynthesis (B) and cholesterol efflux (C). qPCR results are presented as mean ± s.d. (n=3 pools of 30 larvae each), expression levels were normalized to β-actin (bac2) and statistical significance was determined using unpaired two-tailed Student’s t-test compared to the control treatment (*P<0.5,**P<0.01,***P<0.001,****P<0.0001). Black bars represent microarray results and white bars RT-qPCR results. T0901317 and GW3965 treatments are abbreviated as T0 and GW, respectively.
To confirm that the identified genes indeed were regulated by Lxr, we microinjected an lxr morpholino into early zebrafish embryos. The expression of fads2, elovl2, hmgcra, and cyp51 genes were downregulated in embryos injected with the lxr morpholino compared to the control morpholino (Supplemental Fig. S2A), indicating that these genes are Lxr-regulated either directly or indirectly.
In order to obtain insights into the transcriptional regulators likely mediating the gene expression changes observed in zebrafish treated with T0 and GW, enriched sub-network analysis was performed with the gene set affected by both ligands. As expected, Lxr was the major transcriptional regulator estimated to be responsible for the gene expression profile resulting from exposure to the ligands (Supplemental Table S3). Additionally, Srebf1 was among the top enriched transcriptional regulators, possibly reflecting that some of the downstream effects of Lxr activation in zebrafish are mediated through increased expression of srebf1. The graphic representation of genes that are known mammalian expression targets of LXR that were co-regulated by T0 and GW in zebrafish is shown in Supplemental Fig. 3.
3.3. Expression of genes associated with visual perception is altered in zebrafish larvae exposed to the Lxr ligands
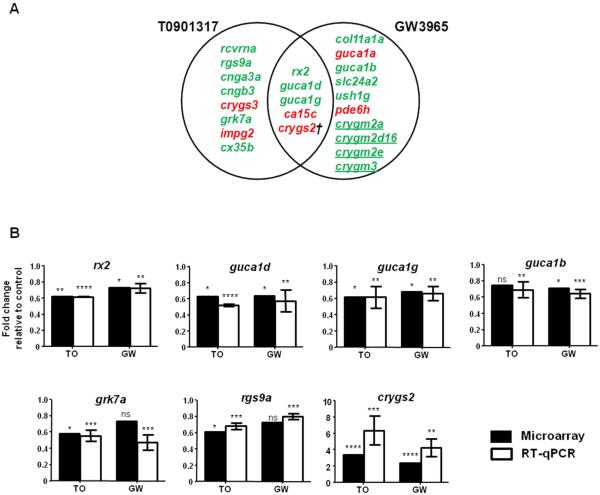
Next, we analyzed the biological functions clustered from the whole set of genes whose expression was regulated by T0 or GW treatment and, as seen in Supplemental Tables 6 and 7, the top enriched biological processes included several lipid molecular clusters. Interestingly, GO enrichment analysis also predicted visual perception as a significantly affected process in zebrafish larvae exposed to either Lxr ligand. Among the visual perception genes whose expression was regulated by T0 and GW, microarray predicted that a subset of 5 genes (rx2, guca1d, guca1g, ca15c, crygs2) was co-regulated by both ligands, while the expression of 8 genes (rcvrna, rgs9, cnga3a, cngb3, crygs3, grk7a, impg2, cx35b) was predicted to be altered by T0, and 10 genes (col11a1a, guca1a, guca1b, slc24a2, ush1g, pde6h, crygm2a, crygm2d16, crygm2e, crygm3) were predicted to be affected by GW (Fig. 3A).
Figure 3. Lxr ligands regulate the expression of genes associated with visual perception in zebrafish larvae.
Venn overlap illustrating visual perception genes regulated by T0901317 and GW3965 in 6 dpf zebrafish larvae (A). Microarray results and RT-qPCR confirmation of selected genes (rx2, guca1d, guca1g, guca1b, grk7a, rgs9a, crygs2) involved in visual function regulated by the Lxr ligands (B). RT-qPCR results are presented as mean ± s.d. (n=3-4 pools of 30 larvae each) and statistical significance was determined using unpaired two-tailed Student’s t-test compared to the control treatment (*P<0.5, **P<0.01, ***P<0.001, ****P<0.0001). Expression levels were normalized to β-actin (bac2). Black bars represent microarray results and white bars RT-qPCR results. T0901317 and GW3965 treatments are abbreviated as T0 and GW, respectively. Upregulated genes in panel A are displayed in red; downregulated genes in green. † indicates that the human orthologue of crygs2 has not been annotated in visual perception GO category. Underlined genes indicate lens gamma crystallins restricted to aquatic vertebrates.
For subsequent RT-qPCR confirmation of genes associated with the visual perception process, we selected genes that the microarray indicated had regulated expression by both T0 and GW (rx2, guca1d, guca1g, crygs2) and by each ligand alone (guca1b, grk7a, rgs9a). The expression of the zebrafish retinal homeobox gene rx2, which has a critical role in eye development, was downregulated in zebrafish larvae exposed to both Lxr ligands. Several genes expressed in zebrafish photoreceptors and directly involved in phototransduction were repressed by treatment with the Lxr ligands, such as the photoreceptor guanylate cyclase activators guca1d, guca1g and guca1b (Fig. 3B). The expression of some visual perception genes were significantly regulated by one LXR ligand in the microarray, and close to significantly regulated by the other ligand, e.g. grk7a (P=0.012 and P=0.094 for T0 and GW, respectively), rgs9a (P=0.019 and P=0.081 for T0 and GW, respectively) or guca1b (P=0.062 and P=0.014 for T0 and GW, respectively). These genes were significantly repressed by both ligands based on RT-qPCR analyses (Fig. 3B), likely reflecting that microarray analysis has been shown not to identify the complete set of genes regulated by a certain ligand (21).
Besides retinal genes, the microarray analysis showed that the expression of several crystallins, major structural proteins of the lens, was regulated by the Lxr ligands. Crystallin gamma S2 (crygs2) expression was induced by both T0 and GW (Fig. 3A, B), crystallin gamma S3 (crygs3) expression was upregulated by T0, while the expression of several gamma M crystallins (crygm2a, crygm2d16, crygm2e, crygm3), which are restricted to aquatic vertebrates, were repressed by GW (Fig. 3A).
In lxr morpholino injected embryos, the expression of grk7a, guca1b, guca1d and guca1g was downregulated compared to control morpholino injected embryos, confirming that these eye specific genes are regulated by Lxr (Supplemental Fig. S2B).
3.4. Lxr ligands regulate genes expressed in liver, yolk syncytial layer and eye
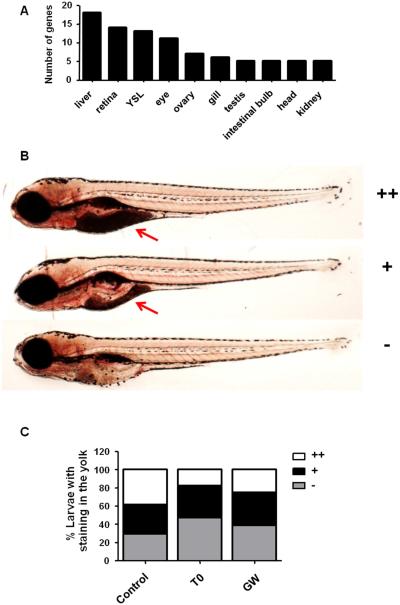
An advantage of using whole zebrafish embryos for transcriptomic analysis is that the tissues that are most affected by a treatment can be mapped. The genes with co-regulated expression by T0 and GW were uploaded to the NIH DAVID Bioinformatic Resources database (22), followed by tissue enrichment analysis using the ZFIN_Anatomy functional category, which is based on published tissue-specific gene expression information from ZFIN database. Out of the 170 genes co-regulated by T0 and GW, 122 were mapped to their respective DAVID IDs, and a total of 43 genes were enriched in the ZFIN_Anatomy functional category. The liver, yolk syncytial layer (YSL) and eye/retina were predicted to be significantly affected tissues in the exposed zebrafish (Fig. 4A, Table S8). This tissue clustering is in accordance with the GO clustering, showing that the expression of genes involved in lipid metabolism and visual perception was strongly regulated by the LXR ligands.
Figure 4. Increased yolk consumption in zebrafish larvae exposed to the Lxr ligands.
Tissue enrichment analysis predicted the liver, retina/eye and yolk syncytial layer (YSL) to be affected by exposure of zebrafish larvae to the Lxr ligands (A). Zebrafish larvae were scored for Oil Red O (ORO) staining in the yolk region from no staining (−), mild (+) to strong (++) staining (B). Number of zebrafish larvae exposed from 4 dpf to 6 dpf to 0.1% DMSO (vehicle), 2 μM T0 and 1 μM GW presenting different staining intensities in the yolk (n=150 larvae for control, n=148 for T0 and n=150 for GW) (C). Red arrow indicates the yolk.
3.5. Increased lipid uptake from the yolk in zebrafish exposed to the Lxr ligands
During the first days of development, zebrafish relies entirely on its yolk sac for the nutrients needed to sustain normal growth, such as triacylglycerol, cholesterol and essential fat-soluble vitamins. The YSL transports nutrients from the yolk to the developing embryo and several genes coding for enzymes involved in early metabolism and nutrient-related functions are expressed within the YSL (23). Considering that lipid metabolism is a major biological process regulated by Lxr in zebrafish, that the yolk is a lipid-rich tissue crucial for early development and that the YSL was predicted to be strongly affected in zebrafish exposed to the Lxr ligands, we assessed whether any changes in lipid staining of the yolk were observed between treatment and control groups using Oil Red O (ORO) staining (Fig. 4B-C). Each larva was scored for intensity in ORO staining ranging from no staining (−), meaning that all yolk was consumed, to strong staining (++), which indicates a very low yolk consumption by the zebrafish larvae (Fig. 4B). We observed that the yolk sac region of the treated groups had less ORO staining in comparison to the control larvae (Figs. 4C), suggesting that exposure to the ligands promoted higher lipid mobilization from the yolk to the developing larvae. Thus, Lxr may play a role in regulating lipid uptake from the yolk during zebrafish development.
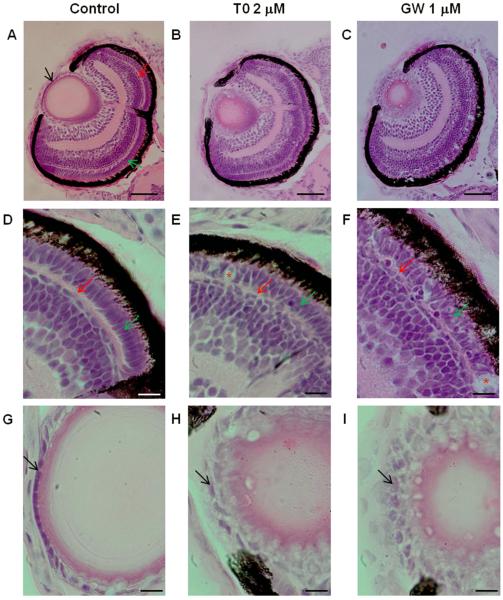
3.6. Exposure to Lxr ligands affects zebrafish eye development
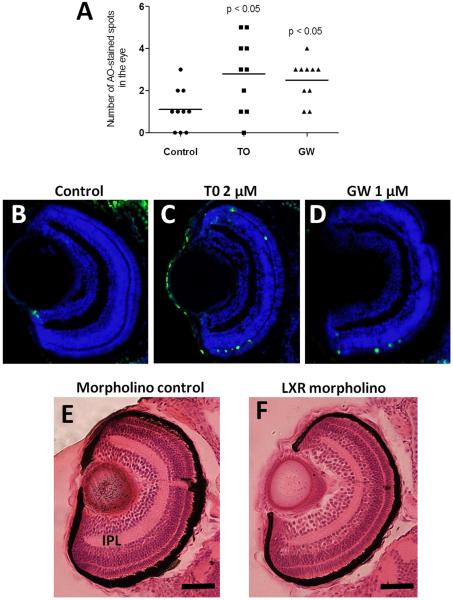
As visual perception was an enriched functional category in zebrafish larvae exposed to T0 and GW, and tissue enrichment analysis predicted the zebrafish eye to be affected by exposure to the Lxr ligands, we assessed whether treatment with the compounds induced morphological changes in the developing zebrafish eye. Zebrafish embryos were treated with 2 μM T0 and 1 μM GW from 6 hpf, before eye field specification, to 4 dpf with daily treatment renewal, and eye sections were stained with H&E. We observed that treatment with the Lxr ligands induced irregularities in the lens and retina of the developing zebrafish eye (Fig. 5). The retinal outer plexiform and photoreceptor layers were disorganized, with indications of photoreceptor cell death (Fig. 5D-F), and the lens epithelial cell layer showed abnormal arrangement and altered epithelial cell morphology (Fig. 5G-I). The increased cell death in GW and TO treated larvae was confirmed by acridine orange staining (Fig. 6A) and was a result of increased apoptosis as determined by TUNEL staining, in particular in the outer nuclear and plexiform layers (Fig. 6B-D).
Figure 5. Morphological alterations in the eye of zebrafish larvae exposed to the LXR ligands.
Hematoxylin and eosin staining of eye sections of zebrafish larvae exposed to vehicle only (0.1 % DMSO), 2 μM T0 and 1 μM GW from 6 hpf to 4 dpf, showing abnormalities in the lens and retina (A-I). Black arrow indicates lens epithelium, red arrow indicates outer plexiform layer and green arrow indicates photoreceptor layer. Disorganized outer plexiform and photoreceptor layers with indications of photoreceptor cell death (indicated by red asterisks) were observed in zebrafish exposed to T0 (B, E) and GW (C, F) compared to control (A, D). The lens epithelial cell layer shows abnormal arrangement and morphology in zebrafish larvae treated with T0 (B, H) and GW (C, I). (Scale bars: A-C: 50 μm: D-I: 10 μm).
Figure 6. Apoptosis and morphological alterations in the eyes of LXR ligand treated or LXR morpholino injected zebrafish larvae.
Number of acridine orange stained spots in eyes of 4 dpf zebrafish larvae treated with T0 and GW (A). TUNEL staining of eye sections of zebrafish larvae treated vehicle only (B), T0 (C) or GW (D), Hematoxylin and eosin staining of eye sections of control morpholino (E) and Lxr morpholino (F) zebrafish larvae at 4 dpf. Representative images are shown. IPL; inner plexiform layer (Scale bars 50 μm).
In LXR morpholino injected fish, eye morphology was also altered. The inner plexiform layer was thinner in the eyes of LXR morpholino injected embryos compared to control injected embryos (Fig. 6E-F). These findings confirm that Lxr is important for normal eye development in zebrafish.
3.7. Lxr ligand exposure induces genes involved in lipid synthesis and visual perception in juvenile zebrafish
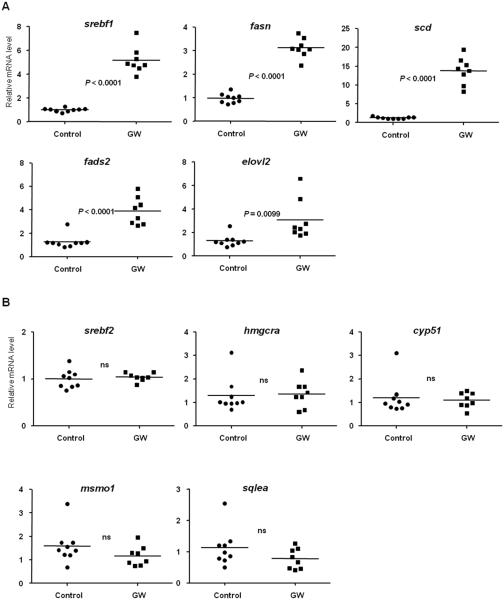
Zebrafish reach adult stage at 3 months. In order to assess if Lxr also regulates the expression of genes associated with lipid biosynthesis and visual perception at later stages during zebrafish development, we exposed 1 month old juvenile zebrafish daily to 1 μM GW for 4 days, and assessed by RT-qPCR whether the treatment induced expression changes in genes related to the major biological processes described to be affected in zebrafish larvae at 6 dpf. Similarly to the results observed in zebrafish larvae, the expression of several genes important for the fatty acid biosynthetic process, such as srebf1, fasn, fads2, scd and elovl2 were upregulated in the treated fish (Fig. 7A). On the contrary, the expression of genes that are involved in cholesterogenesis was not affected by treatment with GW in juvenile zebrafish (Fig. 7B). Similar results were obtained for repeated daily exposures to 1 μM T0 in juvenile fish (Supplemental Fig. S4).
Figure 7. Regulation of genes involved in lipid metabolism in juvenile zebrafish treated with GW.
1 month old juvenile zebrafish were treated with 1 μM GW or vehicle only (0.1% DMSO) daily for 4 days, and the mRNA content of the genes related to fatty acid synthesis srebf1, fasn, scd, fads2 and elovl2 (A) and cholesterol biosynthesis srebf2, hmgcra, cyp51, msmo1 and sqlea (B) was assessed by RT-qPCR. Expression levels were normalized to β-actin (bac2) and each individual value is presented with the mean indicated by a line; Control (solid circles), n=9 and GW (solid squares), n=8. Statistical analysis was performed with unpaired two-tailed Student’s t-test.
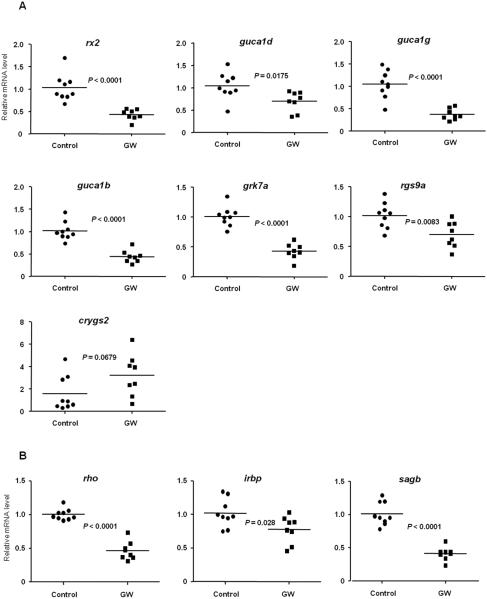
Moreover, the expression of genes involved in visual perception that was affected in the 6 dpf larvae (rx2, guca1d, guca1g, guca1b, grk7a, rgs9a, crygs2) was regulated similarly in the juvenile fish treated with GW, demonstrating that exposure to Lxr ligands also target the eye and genes important for visual perception at later developmental stages in zebrafish (Fig. 8A).
Figure 8. Regulation of genes involved in visual perception in juvenile zebrafish treated with GW.
1 month old juvenile zebrafish were treated with 1 μM GW or vehicle only (0.1% DMSO) daily for 4 days, and the mRNA content of the genes related to visual perception rx2, guca1d, guca1g, guca1b, grk7a, rgs9a and crygs2 was assessed by RT-qPCR (A). 1 month old juvenile zebrafish were treated with 1 μM GW or vehicle only (0.1% DMSO) daily for 4 days, and the mRNA level of genes containing the PCE-1 conserved element (rho, irbp, sagb) was assessed by RT-qPCR (B). Expression levels were normalized to β-actin (bac2) and each individual value is presented with the mean indicated by a line; Control (solid circles), n=9 and GW (solid squares), n=8. Statistical analysis was performed with unpaired two-tailed Student’s t-test.
As previously shown, expression of retinal homeobox gene 2 (rx2) was affected by treatment with Lxr ligands at both larval and juvenile stages in zebrafish. Retinal homeobox gene products have been shown to bind to a conserved sequence element known as Photoreceptor Conserved Element 1 (PCE-1 or Ret-1), found in the promoters of several genes expressed in photoreceptors, including rhodopsin (rho), red cone opsin, rod arrestin (sagb), and interphotoreceptor retinoid-binding protein (irbp) genes in Xenopus (24). Since rx2 expression was downregulated by treatment with the Lxr agonists in zebrafish, we investigated whether the expression levels of genes under the control of PCE-1 regulatory regions would be similarly repressed in zebrafish. As shown in Figure 8B, the expression of the genes rho, sagb and irbp was also significantly downregulated by GW treatment in juvenile zebrafish. These results suggest that Lxr signaling could be involved in a network that modulates expression of retinal homeobox and its downstream genes in zebrafish.
4. Discussion
4.1. Effects of Lxr activation on cholesterol transport and lipid metabolism in zebrafish
Several studies demonstrate that LXRs have key physiological functions in the control of cholesterol transport and homeostasis in mammals (1). LXRs protect against cholesterol overload by directly upregulating the expression of the ATP-binding cassette cholesterol transporters Abca1 and Abcg1, which are involved in reverse cholesterol transport from peripheral tissues to the liver. Additionally, LXR activation stimulates hepatic cholesterol excretion by inducing the transcription of Cyp7a1 in mice, the rate limiting step enzyme in bile acid synthesis, and by stimulating the expression of the cholesterol transporters Abcg5 and Abcg8 in the liver, where they drive cholesterol efflux into the bile. In the intestine, LXR inhibits absorption of dietary cholesterol by upregulating the expression of abcg5 and abcg8 in enterocytes. In zebrafish, Lxr overexpression in intestinal enterocytes delays lipid absorption and promotes intestinal neutral lipid storage (25).
We show here that the ATP-binding cassette transporters abca1a, abcg1, abcg5 and abcg8 are regulated by Lxr ligands in zebrafish larvae. This is in accordance with previous studies that have shown that pharmacological activation of Lxr induces expression of classical Lxr downstream genes, such as abca1a and abcg1 in zebrafish embryos (13) and in adult zebrafish liver (14). Moreover, the expression of abcg8 is lower in liver of adult Lxr knock out fish than of wt fish (25). In addition, several other genes involved in cholesterol transport/homeostasis (e.g. abcg4, apoa4a, ldlr) and in plasma lipoprotein transport/metabolism (e.g. pltp, lpl) was regulated in zebrafish larvae.
In addition to cholesterol transport and homeostasis, LXRs are known to regulate hepatic fatty acid (FA) synthesis in mammalian models by stimulating the transcription of sterol regulatory binding factor 1 (Srebf1), a master regulator of lipogenesis, and genes involved in de novo synthesis and desaturation of fatty acids, such as acetyl-CoA carboxylase (Acaca), the rate limiting step in FA biosynthesis, fatty acid synthase (Fasn) and stearoyl-CoA desaturase (Scd). Our transcriptomic analysis revealed that enzymes important for de novo synthesis and desaturation of FAs, such as acaca, fasn and scd were upregulated in the developing larvae treated with both T0 and GW. This is also in accordance with previous zebrafish studies (14, 15).
Additionally, our study demonstrated that expression of genes related to long chain polyunsaturated fatty acid (LC-PUFA) synthesis was regulated after exposure of zebrafish larvae to T0 and GW. LC-PUFAs are incorporated in phospholipids and are critical for the development of neuronal tissues (26). In zebrafish, the fatty acyl desaturase fads2, and the elongase of very long fatty acids elovl2, are highly expressed in the head region and YSL of 1 day old zebrafish embryos, suggesting that these genes have an important role in brain development and remodeling of yolk fatty acids during zebrafish early embryogenesis (26). In our microarray results, fads2 and elovl2 were among the top ranked genes with upregulated expression in zebrafish larvae exposed to the Lxr ligands, and were significantly downregulated in the lxr morpholino injected fish, confirming a role of Lxr in the control of LC-PUFA synthesis in the developing zebrafish A schematic representation of all the fatty acid biosynthetic genes identified as targets of Lxr activation in zebrafish according to our microarray data is shown in Supplemental Figure S5.
In mammals, LXR regulation of lipogenesis occurs either through a direct activation of genes associated with lipid biosynthesis or in an SREBF1-dependent manner (27, 28). While liganded Lxr has been shown to activate srebf1 in a cell line derived from Atlantic salmon (29), we describe here in vivo that Lxr mediated the induction of srebf1, suggesting that the function of srebf1 as a key metabolic regulator of LXR-mediated signaling is conserved between mammals and fish.
Lxr also activated expression of srebf2, which directly regulates several of the enzymes responsible for cholesterol synthesis (30, 31), in zebrafish embryos. Accordingly, the expression of several enzymes associated with cholesterol synthesis was upregulated in zebrafish larvae exposed to T0 and GW (Fig. 2B and Supplemental Fig. S6). Expression of hmgcra and cyp51 was repressed in lxr morpholino-injected fish. These findings indicate a function of Lxr in regulating, either directly or indirectly, cholesterol biosynthesis during early zebrafish development. Interestingly, expression of srebf2 and several of its target genes was not regulated in juvenile zebrafish daily treated with GW for 4 days. We can also speculate that at very early developmental stages, pharmacological systemic Lxr activation decreased cholesterol in the larvae to levels that are inadequate for proper development, and in turn a feedback mechanism to synthesize cholesterol de novo is activated. This effect on cholesterogenesis may not have been induced in juvenile fish due to external feeding regimen and/or mobilization of cholesterol esters from white adipose tissue, which is formed in zebrafish at 12 dpf (32).
During early development, zebrafish relies completely on its yolk sac for lipids and nutrients needed to sustain growth and survival, and by 5-6 dpf, the yolk is depleted and zebrafish larvae must acquire lipids from feed (33). Zebrafish tissue enrichment analysis predicted the yolk syncytial layer (YSL) as a major tissue affected in zebrafish larvae in response to exposure to the Lxr ligands. The YSL is a transient extra-embryonic tissue that is crucial for the transport of nutrients from the yolk to the zebrafish embryo during early development, and several important genes for lipid remodeling and uptake are expressed in the YSL (23). We observed that zebrafish larvae treated with the Lxr agonists displayed less Oil Red O staining in the yolk region compared to the control larvae, suggesting that Lxr induces the mobilization and utilization of lipids from the yolk during zebrafish development. Accordingly, Lxr knockdown by morpholino resulted in reduced yolk utilization in zebrafish embryos, evidenced by higher levels of cholesterol in the yolk of Lxr morphants compared to controls (13). In mammals, both LXRs are detected in the yolk sac and placenta during fetal development, and coexpression of Rxrα, Lxrα and Abca1 has been established in mouse yolk sac membranes and placental structures (34). Furthermore, treatment of pregnant mice with T0 upregulated Abca1 expression in placental tissue and stimulated maternal-fetal cholesterol transport, suggesting that LXRs may be implicated in metabolism and/or transport of lipids from extra-embryonic tissues to the developing fetus during mammalian gestation (35).
4.2. Effects of Lxr activation in the zebrafish eye
We have recently shown that lxr is expressed in the neural retina of developing and adult zebrafish, and pharmacological activation of Lxr affects the expression of genes related to lipid metabolism and cholesterol transport in the adult zebrafish eye (13). Here, we show that Lxr activation in larval and juvenile zebrafish can also impact the transcription of several genes that are expressed in the eye and directly involved in visual perception and eye development. These genes included the Retinal homeobox gene, rx2, which encodes a key transcription factor for the organogenesis of the vertebrate eye. Rx2 is important for the specification of the retinal field during eye development (36, 37). Consequently, overexpression of Rx2 results in the loss of forebrain tissue and the ectopic formation of retinal tissue. In the adult retina rx2 is expressed exclusively in cones, but not rods. In addition, several other genes expressed in photoreceptors and directly involved in the photoresponse process had a downregulated expression by exposure to Lxr ligands, including guanylate cyclase-activating proteins (guca1b, guca1d, guca1g), which are Ca2+-binding proteins of the calmodulin gene superfamily that function as regulators of photoreceptor guanylate cyclases. In zebrafish retina, guca1d (gcap4) and guca1g (gcap7) are expressed in cone photoreceptors, while guca1b (gcap2) is detected in both cones and rods (38, 39). Expression of the cone-specific kinase grk7a, important for cone pigment phosphorylation and normal cone response recovery in zebrafish (40), was also downregulated upon exposure to the LXR ligands. These retinal genes (guca1b, guca1d, guca1g and grk7a) were similarly affected in the lxr morpholino injected fish, indicating a function of lxr in regulating the expression of genes important for phototransduction in zebrafish. Additionally, Lxr activation resulted in repression of the expression of regulator of G-protein signaling 9a (rgs9a), responsible for the inactivation of the G protein transducin during photoresponse recovery from excitation (41), and several other genes that encode functional components of the phototransduction cascade, such as the nucleotide-gated cation channels cnga3a and cngb3 (42) and the γ subunit of the cGMP-specific phosphodiesterase complex (pde6h) present in cone photoreceptor outer segments. In the juvenile zebrafish treated with the GW, the expression of rx2 and its putative downstream genes rho, sag and irbp was downregulated, suggesting that Lxr signaling could be involved in the modulation of rx activity and play a role in photoreceptor-specific gene expression in zebrafish at later developmental stages.
Lxr is also detected in the lenticular tissue in the developing zebrafish eye (13). In accordance, the expression of several crystallins, major structural and protective proteins in the lens, were altered by the Lxr ligands, including crygs2, crygm2a, crygm2d16, crygm2e and crygm3.
In both ligand treated embryos and lxr morpholino embryos, morphological alterations were identified in the eyes. The molecular mechanisms underlying these abnormalities, as well as the transcriptional regulation of photoreceptor-specific genes and crystallins need to be further investigated for better understanding of the role of Lxr in the eye. Noteworthy, sterol-27-hydroxylase (Cyp27a1) and cholesterol-24-hydroxylase (Cyp46a1), enzymes responsible for the synthesis of 27-hydroxycholesterol and 24(S)-hydroxycholesterol, respectively, are expressed in mammalian retina (43, 44), and these oxysterols may have functional roles as endogenous LXR ligands in normal retinal development and physiology.
5. Conclusions
In conclusion, our data demonstrate a primary role of Lxr in regulating lipid metabolic processes during zebrafish development. Several LXR-mediated transcriptional effects observed in mammalian systems were conserved in zebrafish, supporting the use of zebrafish as an alternative in vivo model for Lxr-related studies involving lipid metabolism and cholesterol homeostasis. Moreover, pharmacological Lxr activation and morpholino knockdown studies indicated that Lxr controls the expression of genes associated with visual perception, and included retinal and lens-specific genes, suggesting a role of Lxr in regulating important components of visual function in zebrafish.
Supplementary Material
Table S1. List of genes with co-regulated expression by T091317 and GW3965. Red genes showed upregulated expression by both ligands, whereas green genes were repressed by both ligands. Blue genes were upregulated by T0901317 and repressed by GW3965. (Fold change ≥1.4; P≤0.05).
Table S5. Top ranked transcriptional regulators of genes co-regulated by T0 and GW in zebrafish larvae.
Table S6. Enriched biological processes obtained from the whole set of differentially expressed genes in zebrafish larvae exposed to T0.
Table S7. Enriched biological processes obtained from the whole set of differentially expressed genes in zebrafish larvae exposed to GW.
Table S8. Enrichment analysis of zebrafish tissues most affected by treatment with T0 and GW (ZFIN_Anatomy NIH DAVID Functional Annotation). Tissue enrichment analysis was performed with the set of co-regulated genes by T0 and GW. Enriched tissues with P<0.05 and with a minimum of 5 enriched genes are represented (YSL = yolk syncytial layer).
Figure S1. GW3965 is a ligand for zfPparγ. Transactivation assay performed in HG5LN cells stably transfected with chimeric GAL4-zfPparγLBD showing that GW3965 activates zfPparγ in vitro, while T0901317 has no effect on zfPparγ-mediated transcriptional activation. Results are presented as mean ± s.d. of 2 independent experiments performed in quadruplicate.
Figure S2. Regulation of expression of Lxr target genes is altered in Lxr morpholino knock-down fish. qPCR analysis was performed on selected genes involved in fatty acid and cholesterol biosynthesis (A), and visual perception (B). qPCR results are presented as mean ± s.d. (n=4 pools of 30 larvae each), expression levels were normalized to β-actin (bac2) and statistical significance was determined using unpaired two-tailed Student’s t-test compared to the control treatment (*P<0.5,**P<0.01,***P<0.001,****P<0.0001).
Figure S3. Graphic representation of expression targets of LXR that were significantly regulated (P≤0.05, fold change ≥|±1.4|) in zebrafish larvae exposed to T0 and GW. Colors reflect direction of expression change (red: upregulation, green: downregulation). Blue arrows indicate expression relationship. Arrows with a “+” in a circle indicate a positive relation. Dead-head arrows (−|) indicate a negative relation. Oval shapes represent protein, circle on tripod platform – transcription factor; ice cream cone – receptor.
Figure S4. Regulation of genes involved in lipid metabolism in zebrafish treated with T0. Juvenile zebrafish were treated with 1 μM T0 or vehicle only (0.1% DMSO) daily for 4 days, and the mRNA content of the genes related to fatty acid synthesis srebf1, fasn, and scd, and cholesterol biosynthesis srebf2, hmgcra, and cyp5,was assessed by RT-qPCR. Expression levels were normalized to β-actin (bac2) and each individual value is presented with the mean indicated by a line; Control (solid circles), n=8 and T0 (solid squares), n=9. Statistical analysis was performed with unpaired two-tailed Student’s t-test.
Figure S5. Schematic representation of the fatty acid (FA) biosynthetic pathway with genes that were upregulated by T0 and GW in zebrafish larvae. Genes that encode enzymes associated with the synthesis of FAs de novo (A) and long-chain polyunsaturated FAs (B) were upregulated by the ligands as indicated by the red arrows. fads2 has dual Δ5- and Δ6- desaturase activity in zebrafish (Monroig et al. 2009). Figure adapted from (45).
Figure S6. Schematic representation of cholesterol biosynthetic pathway indicating genes upregulated by T0 and GW in zebrafish larvae. Expression of several genes that encode enzymes associated with cholesterol biosynthesis was significantly upregulated by the LXR ligands as indicated by the red arrows. Green arrows represents genes only significantly upregulated by T0. Figure adapted from (45).
Table S2. List of genes with altered expression by T0 only. Red and green genes showed upregulated and downregulated expression, respectively, in zebrafish larvae exposed to T0 (Fold change ≥1.4; P≤0.05).
Table S3. List of genes with altered expression by GW only. Red and green genes showed upregulated and downregulated expression, respectively, in zebrafish larvae exposed to GW (Fold change ≥1.4; P≤0.05).
Table S4. Primer sequences used for the RT-qPCR validation of genes from the microarray.
Highlights.
We performed a microarray analysis of Lxr regulated gene expression in zebrafish
Lxr regulated expression of genes involved in lipid metabolic processes
Lxr ligands induced changes in expression of genes in the neural retina and lens
Lxr activation increased zebrafish embryo yolk absorption
Lxr activation caused morphological alterations in the retina and lens of the zebrafish eye
6. Acknowledgements
We thank Drs. Wanfu Wu and Yu-Bing Dai for advice on sectioning and staining. This study was funded by grants from the Environmental Protection Agency (R834289) and the National Institute of Health/National Institute of Environmental Health Sciences (R21ES020036), and the Robert A. Welch Foundation (E-0004). The views expressed in the article do not necessarily reflect the views of the funders.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- 1.Baranowski M. Biological role of liver X receptors. J Physiol Pharmacol. 2008;59(Suppl 7):31–55. [PubMed] [Google Scholar]
- 2.Teboul M, Enmark E, Li Q, Wikstrom AC, Pelto-Huikko M, et al. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc Natl Acad Sci U S A. 1995;92:2096–100. doi: 10.1073/pnas.92.6.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jakobsson T, Treuter E, Gustafsson JA, Steffensen KR. Liver X receptor biology and pharmacology: new pathways, challenges and opportunities. Trends Pharmacol Sci. 2012;33:394–404. doi: 10.1016/j.tips.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Gabbi C, Warner M, Gustafsson JA. Minireview: liver X receptor beta: emerging roles in physiology and diseases. Mol Endocrinol. 2009;23:129–36. doi: 10.1210/me.2008-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steffensen KR, Jakobsson T, Gustafsson JA. Targeting liver X receptors in inflammation. Expert Opin Ther Targets. 2013;17:977–90. doi: 10.1517/14728222.2013.806490. [DOI] [PubMed] [Google Scholar]
- 6.Krasowski MD, Ai N, Hagey LR, Kollitz EM, Kullman SW, et al. The evolution of farnesoid X, vitamin D, and pregnane X receptors: insights from the green-spotted pufferfish (Tetraodon nigriviridis) and other non-mammalian species. BMC Biochem. 2011;12:5. doi: 10.1186/1471-2091-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annicotte JS, Schoonjans K, Auwerx J. Expression of the liver X receptor alpha and beta in embryonic and adult mice. Anat Rec A Discov Mol Cell Evol Biol. 2004;277:312–6. doi: 10.1002/ar.a.20015. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto A, Kawasaki T, Kazawa T, Ohashi R, Jiang S, et al. Expression of liver X receptor alpha in rat fetal tissues at different developmental stages. J Histochem Cytochem. 2007;55:641–9. doi: 10.1369/jhc.6A7120.2007. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, et al. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. 2002;99:13878–83. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacchetti P, Sousa KM, Hall AC, Liste I, Steffensen KR, et al. Liver X receptors and oxysterols promote ventral midbrain neurogenesis in vivo and in human embryonic stem cells. Cell Stem Cell. 2009;5:409–19. doi: 10.1016/j.stem.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Xing Y, Fan X, Ying D. Liver X receptor agonist treatment promotes the migration of granule neurons during cerebellar development. J Neurochem. 2010;115:1486–94. doi: 10.1111/j.1471-4159.2010.07053.x. [DOI] [PubMed] [Google Scholar]
- 12.Fan X, Kim HJ, Bouton D, Warner M, Gustafsson JA. Expression of liver X receptor beta is essential for formation of superficial cortical layers and migration of later-born neurons. Proc Natl Acad Sci U S A. 2008;105:13445–50. doi: 10.1073/pnas.0806974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archer A, Kitambi SS, Hallgren SL, Pedrelli M, Olsen KH, et al. The Liver X-Receptor (Lxr) Governs Lipid Homeostasis in Zebrafish during Development. Open Journal of Endocrine and Metabolic Diseases. 2012;2:74–81. [Google Scholar]
- 14.Archer A, Lauter G, Hauptmann G, Mode A, Gustafsson JA. Transcriptional activity and developmental expression of liver X receptor (lxr) in zebrafish. Dev Dyn. 2008;237:1090–8. doi: 10.1002/dvdy.21476. [DOI] [PubMed] [Google Scholar]
- 15.Sukardi H, Zhang X, Lui EY, Ung CY, Mathavan S, et al. Liver X receptor agonist T0901317 induced liver perturbation in zebrafish: histological, gene set enrichment and expression analyses. Biochim Biophys Acta. 2012;1820:33–43. doi: 10.1016/j.bbagen.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–8. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seimandi M, Lemaire G, Pillon A, Perrin A, Carlavan I, et al. Differential responses of PPARalpha, PPARdelta, and PPARgamma reporter cell lines to selective PPAR synthetic ligands. Anal Biochem. 2005;344:8–15. doi: 10.1016/j.ab.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Riu A, McCollum CW, Pinto CL, Grimaldi M, Hillenweck A, et al. Halogenated bisphenol-A analogs act as obesogens in zebrafish larvae (Danio rerio) Toxicol Sci. 2014;139:48–58. doi: 10.1093/toxsci/kfu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao R, Bondesson M, Singh AV, Riu A, McCollum CW, et al. Identification of Estrogen Target Genes during Zebrafish Embryonic Development through Transcriptomic Analysis. PLoS One. 2013;8:e79020. doi: 10.1371/journal.pone.0079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis MC, et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45:1963–6. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 21.Katchy A, Pinto C, Jonsson P, Nguyen-Vu T, Pandelova M, et al. Co-exposure to Phytoestrogens and Bisphenol A mimic estrogenic effects in an additive manner. Toxicol Sci. 2013 doi: 10.1093/toxsci/kft271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho L, Heisenberg CP. The yolk syncytial layer in early zebrafish development. Trends Cell Biol. 2010;20:586–92. doi: 10.1016/j.tcb.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y, Martinez-De Luna RI, Lou CH, Nekkalapudi S, Kelly LE, et al. Regulation of photoreceptor gene expression by the retinal homeobox (Rx) gene product. Dev Biol. 2010;339:494–506. doi: 10.1016/j.ydbio.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Garcia L, Schlegel A. Lxr-driven enterocyte lipid droplet formation delays transport of ingested lipids. J Lipid Res. 2014;55:1944–58. doi: 10.1194/jlr.M052845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monroig O, Rotllant J, Sanchez E, Cerda-Reverter JM, Tocher DR. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim Biophys Acta. 2009;1791:1093–101. doi: 10.1016/j.bbalip.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshikawa T, Shimano H, Amemiya-Kudo M, Yahagi N, Hasty AH, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmona-Antonanzas G, Tocher DR, Martinez-Rubio L, Leaver MJ. Conservation of lipid metabolic gene transcriptional regulatory networks in fish and mammals. Gene. 2014;534:1–9. doi: 10.1016/j.gene.2013.10.040. [DOI] [PubMed] [Google Scholar]
- 30.Horton JD. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Trans. 2002;30:1091–5. doi: 10.1042/bst0301091. [DOI] [PubMed] [Google Scholar]
- 31.Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, et al. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998;101:2331–9. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imrie D, Sadler KC. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev Dyn. 2010;239:3013–23. doi: 10.1002/dvdy.22443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson JL, Carten JD, Farber SA. Zebrafish lipid metabolism: from mediating early patterning to the metabolism of dietary fat and cholesterol. Methods Cell Biol. 2011;101:111–41. doi: 10.1016/B978-0-12-387036-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marceau G, Volle DH, Gallot D, Mangelsdorf DJ, Sapin V, et al. Placental expression of the nuclear receptors for oxysterols LXRalpha and LXRbeta during mouse and human development. Anat Rec A Discov Mol Cell Evol Biol. 2005;283:175–81. doi: 10.1002/ar.a.20157. [DOI] [PubMed] [Google Scholar]
- 35.Lindegaard ML, Wassif CA, Vaisman B, Amar M, Wasmuth EV, et al. Characterization of placental cholesterol transport: ABCA1 is a potential target for in utero therapy of Smith-Lemli-Opitz syndrome. Hum Mol Genet. 2008;17:3806–13. doi: 10.1093/hmg/ddn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–8. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- 37.Chuang JC, Raymond PA. Zebrafish genes rx1 and rx2 help define the region of forebrain that gives rise to retina. Dev Biol. 2001;231:13–30. doi: 10.1006/dbio.2000.0125. [DOI] [PubMed] [Google Scholar]
- 38.Imanishi Y, Li N, Sokal I, Sowa ME, Lichtarge O, et al. Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur J Neurosci. 2002;15:63–78. doi: 10.1046/j.0953-816x.2001.01835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imanishi Y, Yang L, Sokal I, Filipek S, Palczewski K, et al. Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1-8) in pufferfish (Fugu rubripes) J Mol Evol. 2004;59:204–17. doi: 10.1007/s00239-004-2614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinner O, Makhankov YV, Biehlmaier O, Neuhauss SC. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron. 2005;47:231–42. doi: 10.1016/j.neuron.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 41.He W, Cowan CW, Wensel TG. RGS9, a GTPase accelerator for phototransduction. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 42.Schon C, Biel M, Michalakis S. Gene replacement therapy for retinal CNG channelopathies. Mol Genet Genomics. 2013;288:459–67. doi: 10.1007/s00438-013-0766-4. [DOI] [PubMed] [Google Scholar]
- 43.Javitt NB. Oxysterols: functional significance in fetal development and the maintenance of normal retinal function. Curr Opin Lipidol. 2007;18:283–8. doi: 10.1097/MOL.0b013e328133851e. [DOI] [PubMed] [Google Scholar]
- 44.Heo GY, Bederman I, Mast N, Liao WL, Turko IV, et al. Conversion of 7-ketocholesterol to oxysterol metabolites by recombinant CYP27A1 and retinal pigment epithelial cells. J Lipid Res. 2011;52:1117–27. doi: 10.1194/jlr.M014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of genes with co-regulated expression by T091317 and GW3965. Red genes showed upregulated expression by both ligands, whereas green genes were repressed by both ligands. Blue genes were upregulated by T0901317 and repressed by GW3965. (Fold change ≥1.4; P≤0.05).
Table S5. Top ranked transcriptional regulators of genes co-regulated by T0 and GW in zebrafish larvae.
Table S6. Enriched biological processes obtained from the whole set of differentially expressed genes in zebrafish larvae exposed to T0.
Table S7. Enriched biological processes obtained from the whole set of differentially expressed genes in zebrafish larvae exposed to GW.
Table S8. Enrichment analysis of zebrafish tissues most affected by treatment with T0 and GW (ZFIN_Anatomy NIH DAVID Functional Annotation). Tissue enrichment analysis was performed with the set of co-regulated genes by T0 and GW. Enriched tissues with P<0.05 and with a minimum of 5 enriched genes are represented (YSL = yolk syncytial layer).
Figure S1. GW3965 is a ligand for zfPparγ. Transactivation assay performed in HG5LN cells stably transfected with chimeric GAL4-zfPparγLBD showing that GW3965 activates zfPparγ in vitro, while T0901317 has no effect on zfPparγ-mediated transcriptional activation. Results are presented as mean ± s.d. of 2 independent experiments performed in quadruplicate.
Figure S2. Regulation of expression of Lxr target genes is altered in Lxr morpholino knock-down fish. qPCR analysis was performed on selected genes involved in fatty acid and cholesterol biosynthesis (A), and visual perception (B). qPCR results are presented as mean ± s.d. (n=4 pools of 30 larvae each), expression levels were normalized to β-actin (bac2) and statistical significance was determined using unpaired two-tailed Student’s t-test compared to the control treatment (*P<0.5,**P<0.01,***P<0.001,****P<0.0001).
Figure S3. Graphic representation of expression targets of LXR that were significantly regulated (P≤0.05, fold change ≥|±1.4|) in zebrafish larvae exposed to T0 and GW. Colors reflect direction of expression change (red: upregulation, green: downregulation). Blue arrows indicate expression relationship. Arrows with a “+” in a circle indicate a positive relation. Dead-head arrows (−|) indicate a negative relation. Oval shapes represent protein, circle on tripod platform – transcription factor; ice cream cone – receptor.
Figure S4. Regulation of genes involved in lipid metabolism in zebrafish treated with T0. Juvenile zebrafish were treated with 1 μM T0 or vehicle only (0.1% DMSO) daily for 4 days, and the mRNA content of the genes related to fatty acid synthesis srebf1, fasn, and scd, and cholesterol biosynthesis srebf2, hmgcra, and cyp5,was assessed by RT-qPCR. Expression levels were normalized to β-actin (bac2) and each individual value is presented with the mean indicated by a line; Control (solid circles), n=8 and T0 (solid squares), n=9. Statistical analysis was performed with unpaired two-tailed Student’s t-test.
Figure S5. Schematic representation of the fatty acid (FA) biosynthetic pathway with genes that were upregulated by T0 and GW in zebrafish larvae. Genes that encode enzymes associated with the synthesis of FAs de novo (A) and long-chain polyunsaturated FAs (B) were upregulated by the ligands as indicated by the red arrows. fads2 has dual Δ5- and Δ6- desaturase activity in zebrafish (Monroig et al. 2009). Figure adapted from (45).
Figure S6. Schematic representation of cholesterol biosynthetic pathway indicating genes upregulated by T0 and GW in zebrafish larvae. Expression of several genes that encode enzymes associated with cholesterol biosynthesis was significantly upregulated by the LXR ligands as indicated by the red arrows. Green arrows represents genes only significantly upregulated by T0. Figure adapted from (45).
Table S2. List of genes with altered expression by T0 only. Red and green genes showed upregulated and downregulated expression, respectively, in zebrafish larvae exposed to T0 (Fold change ≥1.4; P≤0.05).
Table S3. List of genes with altered expression by GW only. Red and green genes showed upregulated and downregulated expression, respectively, in zebrafish larvae exposed to GW (Fold change ≥1.4; P≤0.05).
Table S4. Primer sequences used for the RT-qPCR validation of genes from the microarray.