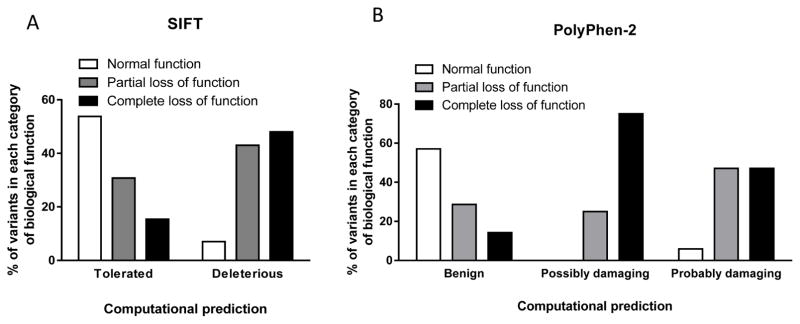
Figure 1. Comparison of computational predictions of the function of GPCR variants by SIFT and PolyPhen-2 with documented biological function test findings.
(A) SIFT and (B) PolyPhen-2 were used to predict the function of 52 variants in GnRHR, KISS1R, PROKR2, and TACR3, for which the results of biological function tests are available. SIFT categorized variants as tolerated or deleterious; PolyPhen-2 categorized variants as benign, possibly damaging, or probably damaging. These predictions were compared with the results of in vitro biological function testing for each variant. Biological function was categorized as normal, partial loss of function, or complete loss of function. There were 10 variants with normal function, 21 with partial loss of function, and 21 with complete loss of function. The percentage of mutations predicted by each program in each category was then calculated.