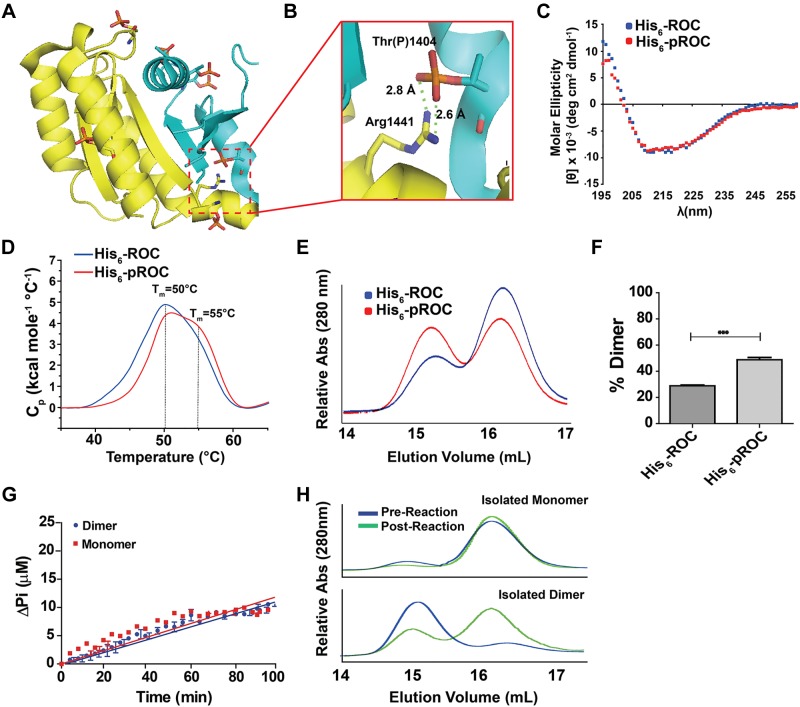
Figure 4.
Phosphorylation of ROC enhances dimer formation. A) Overlay of all phosphorylation occurring in His6-pROC based on ROC crystal structure (PDB ID 2zej). Yellow and cyan represent chain A and B in the dimer, respectively. Atoms on key residues are color coded (phosphorus: orange, oxygen: red, nitrogen: blue, carbon: yellow/cyan). B) Enlargement of the dimerization interface, with the calculated distances indicated. The Thr(P)1404 is predicted to form a salt bridge with the Arg1441 residue. C) CD profiles. No secondary structure differences were detected between His6-ROC and His6-pROC. D) Differential scanning calorimetry profiles. Both His6-ROC and His6-pROC displayed peaks at ∼50°C, whereas the His6-pROC also had a broad shoulder peak with predicted Tm ≅ 55°C. C, D) Traces are representative of at least 3 independent experiments. E) Representative elution profiles of His6-ROC and His6-pROC, immediately after a 3 h kinase reaction (Fig. 1B). E) GTPase assay with purified monomer and dimer His6-ROC. Near equivalent velocities of hydrolyzed GTP at 5.8 ± 0.1 and 6.1 ± 0.2 nmol · min−1 per micromole protein, respectively, were noted. F) Comparative analysis of the proportion of dimer to monomer. Maximum relative peak heights were calculated from 3 independent runs and are displayed as column graphs with bars representing sem. ***P < 0.00; unpaired t test. G) Elution profiles of purified monomer (top) and dimer (bottom) for His6-ROC before (blue) and after (green) the 2 h GTPase assay. Profiles shown are representative of 3 independent runs.