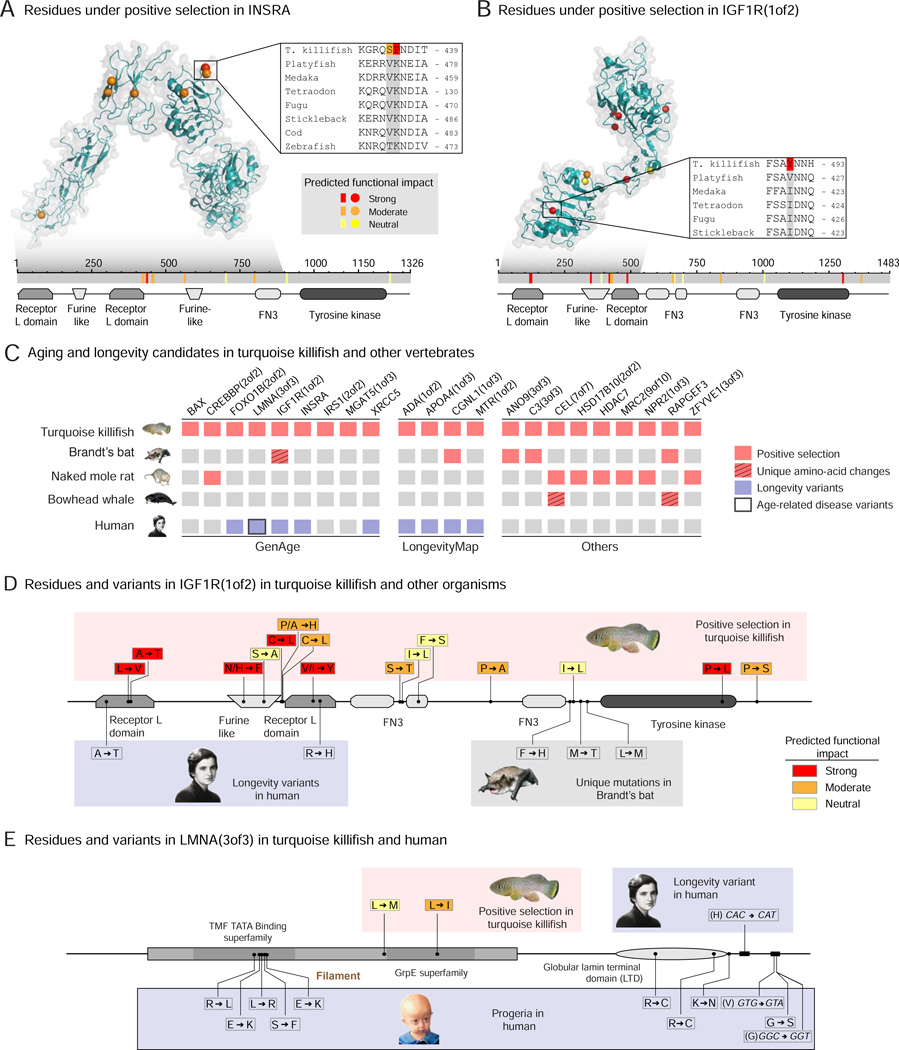
Figure 4. Aging and longevity genes under positive selection in the turquoise killifish and with variants in long-lived species or in humans.
A-B) Location of residues under positive selection and with putative functional consequences in insulin receptor A (INSRA) (A), IGF1R(1of2) (B) in the turquoise killifish. Top panels: crystal structure of human orthologs. Color represents the strength of functional impact. Grey shadow: region of the protein with available crystal structure. Insert: alignment of an example residue with strong functional effect in the turquoise killifish and other fish. Bottom panels: schematic of the residues mapped on the turquoise killifish protein sequence (grey). Colored bars: residues under positive selection with different functional impacts. The conserved protein domains and functional sites are also indicated. FN3: Fibronectin type-III repeats.
(C) Aging and longevity candidates under positive selection in the short-lived turquoise killifish and their variation in long-lived animal species and in humans. Left: aging-related genes from the GenAge database (human and mouse combined, Table S4A). Middle: genes identified in association studies in humans from the LongevityMap database (Table S4A). Right: other genes that are also under positive selection or uniquely changed in other species with extreme longevity phenotype (naked mole rat, Brandt’s bat, bowhead whale).
D) Location and variants of residues under positive selection in the turquoise killifish for IGF1R(1of2), and their location and variants in long-lived species and in human centenarians. Top: turquoise killifish variants, with the changed amino acid on the right. Color represents the strength of functional impact. Bottom: variants associated with centenarians in humans or residues with unique amino acid changes in long-lived Brandt’s bat mapped on the turquoise killifish sequence. The variants correspond to the amino acids on the right.
E) Location and variants of residues under positive selection in the turquoise killifish for LMNA(3of3), and their location and variants in progeria and human centenarians. Top left: turquoise killifish variants, with the changed amino acid on the right. Top right: variants associated with centenarians in humans mapped onto the turquoise killifish sequence. Bottom: Variants in Hutchinson-Gilford Progeria Syndrome mapped onto the turquoise killifish sequence. These variants are the amino acids on the right. For the turquoise killifish, color represents the strength of functional impact.