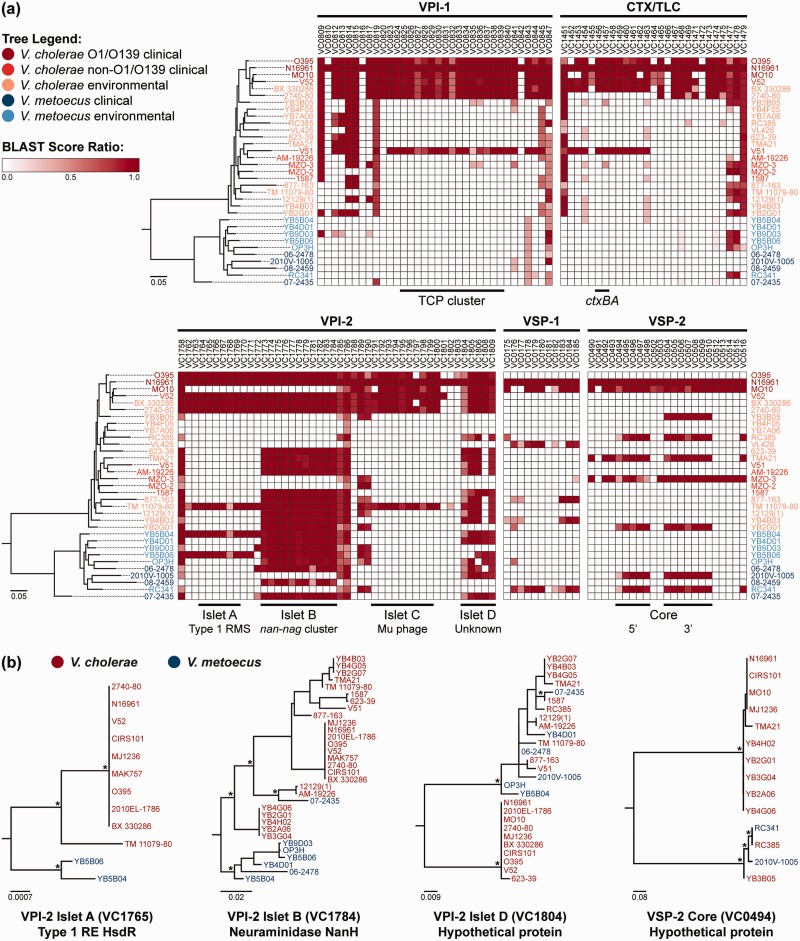
Fig. 5.—
Virulence factors present in V. metoecus and V. cholerae. (a) The phylogenetic relationship of the V. metoecus and V. cholerae strains is shown on the left of each BSR map. The ML phylogenetic tree was constructed using 3,978 amino acid positions based on 400 universally conserved bacterial and archaeal proteins. The scale bars represent amino acid substitutions per site. The columns on the BSR maps show genes (locus tags) from genomic islands VPI-1, CTX/TLC, VPI-2, VSP-1, and VSP-2 of the reference, V. cholerae N16961. The black bars at the bottom of the BSR maps indicate the TCP cluster of VPI-1, ctxAB of CTX/TLC, islets of VPI-2, and core regions of VSP-2. The gradient bar shows the BSRs and their corresponding colors, with white regions indicating the absence of genes. Only BSR values of at least 0.3 were included. VPI, Vibrio pathogenicity island; CTX/TLC, cholera toxin/toxin-linked cryptic; VSP, Vibrio seventh pandemic island; RMS, restriction-modification system. (b) Representative ML phylogenetic trees of orthologous gene families of the VPI-2 islets and the VSP-2 core. Relevant bootstrap support (>70%) is indicated with *. The scale bars represent nucleotide substitutions per site. RE, restriction endonuclease.