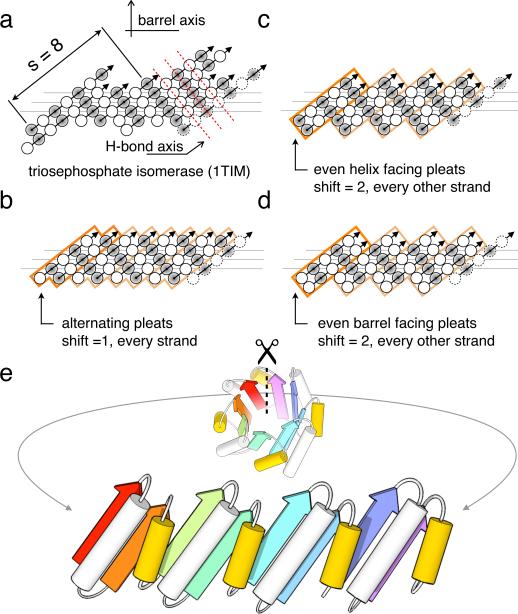
Figure 1. Geometric constraints on the secondary structure arrangement in an ideal 4-fold symmetric TIM-barrel.
(a) The asymmetric β-strand arrangement of the classic triosephosphate isomerase barrel from chicken muscle (PDB code: 1TIM). The strands, as defined by DSSP40, are viewed from the inside of the barrel, with open and shaded circles representing amino acid residues pointing into and out of the barrel, respectively. The first strand is shown at the left and right. Three horizontal lines indicate the residues lining the interior of the barrel. Sheet hydrogen bonds follow diagonal directions indicated by dashed red lines. (b, c, d) The three solutions for achieving the s = 8 shear with 4-fold symmetry. Orange boxes represent 4 individual repeat units; the first unit is highlighted for clarity. Since residues represented by open and shaded circles are structurally non-equivalent and the diagonal running hydrogen bonds must connect residues pointing in the same direction, an 8-fold symmetric barrel is not possible. (b) Register shift of one residue between every strand. (c) Register shift alternating between 0 and 2 residues around the barrel, with strands starting with residues pointing towards the helices (shaded circles). (d) Alternating (0,2) register shift as in c., with strands starting with residues pointing into the barrel (open circles). (e) To achieve the shear pattern in d, the helix spanning the offset strands (yellow) must be shorter than the helix within the repeat unit.