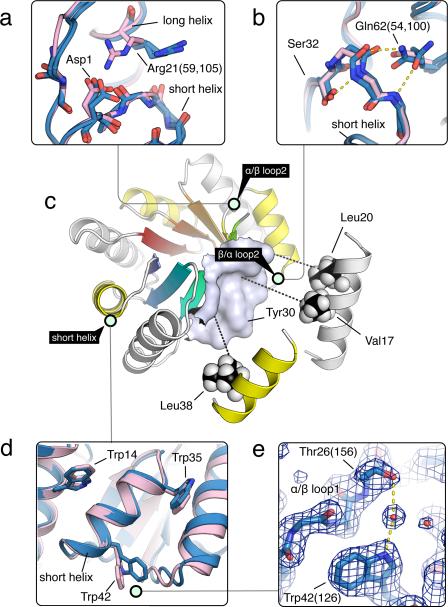
Figure 2. Sequence determinants of de novo designed TIM-barrel.
Designed TIM-barrel model is depicted with light green circles tagging regions shown in the insets, where the design models are shown in pink and X-ray structures in blue. Residues are numbered by the design model, but X-ray structure residue numbers are in parentheses. (a) The α/β loop at the interface between the repeat units with a register shift of 2. Asp1 was designed to satisfy the hydrogen bonding requirement for the backbone amide group on the neighboring strand. In the crystal, Arg21 makes lattice contacts rather than the designed interaction; in solution the designed hydrogen bond may be formed. (b) Features stabilizing the β/α loop backbone. Ser32 was designed to interact with the amide group that points towards the hydrophobic core, and Gln62 with the carbonyl similar to Arg21 (in a). Alternative conformations of Gln62 are observed in two different repeats in the crystal structure. (c) Packing of the sheet facing side of the helices (long helix, white; short helix, yellow) against the surface on the sheet. (d) The wedges between the helices are filled by tryptophans. Trp42 was found to adopt a different conformation in the crystal structure. (e) Trp42 was designed to interact with Thr26 on the neighboring loop directly, but crystallographic evidence suggests that the same interaction is mediated by water, as shown by the clear electron density bridging the two residues.