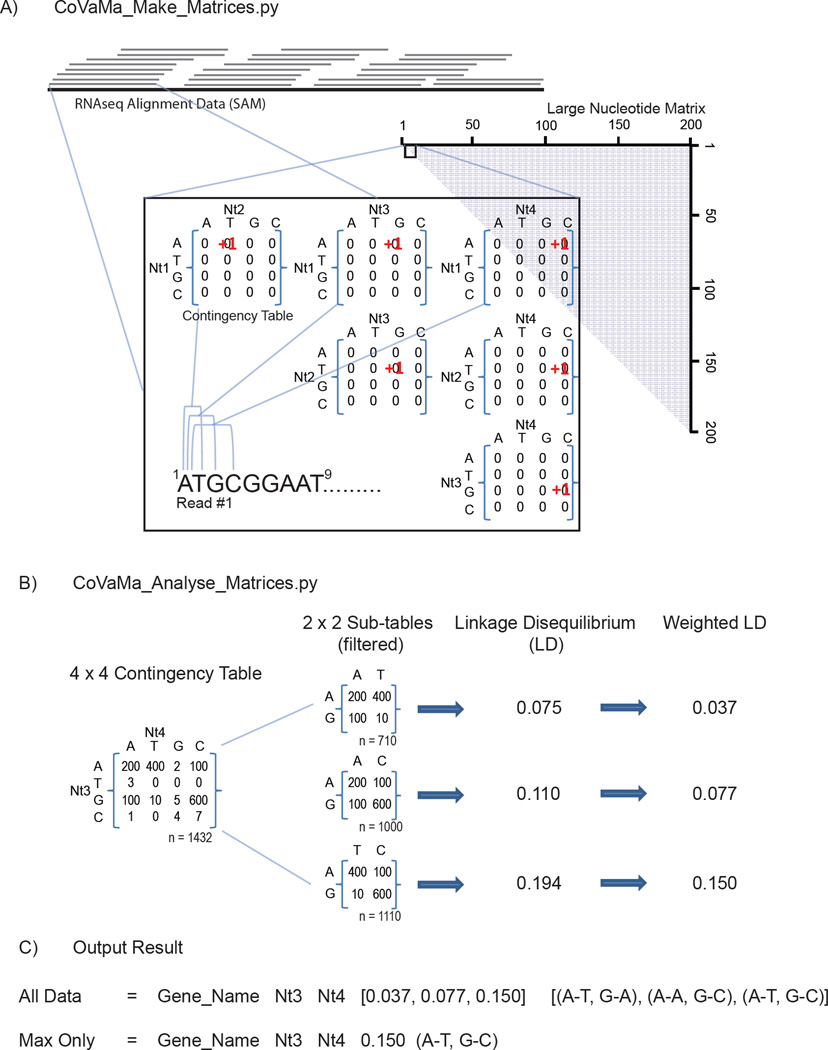
Figure 1.
CoVaMa generates matrices of pairwise nucleotide or amino acid associations from Sequencing Alignment Mapping (SAM) data from RNAseq. A) CoVaMa_Make_Matrices.py is the first script in CoVaMa that extracts information from each individual aligned read. Every possible pairwise interaction of nucleotides in the aligned read is used to populate 4×4 contingency tables that are components of the large nucleotide matrix. Amino acid matrices containing 20×20 contingency tables are built in an analogous fashion, but by first translating each aligned read in the desired reading frame. B) CoVaMa_Analyse_Matrices.py analyses each individual contingency table in the large matrices for evidence of linkage disequilibrium. All possible combinations of 2×2 sub-tables are extracted from the 4×4 (or 20×20) contingency table provided there are sufficient reads mapped. An LD value is calculated for each 2×2 table and optionally normalized relative to the total number of reads in the 4×4 table. Results are written to an output file as exemplified in C).