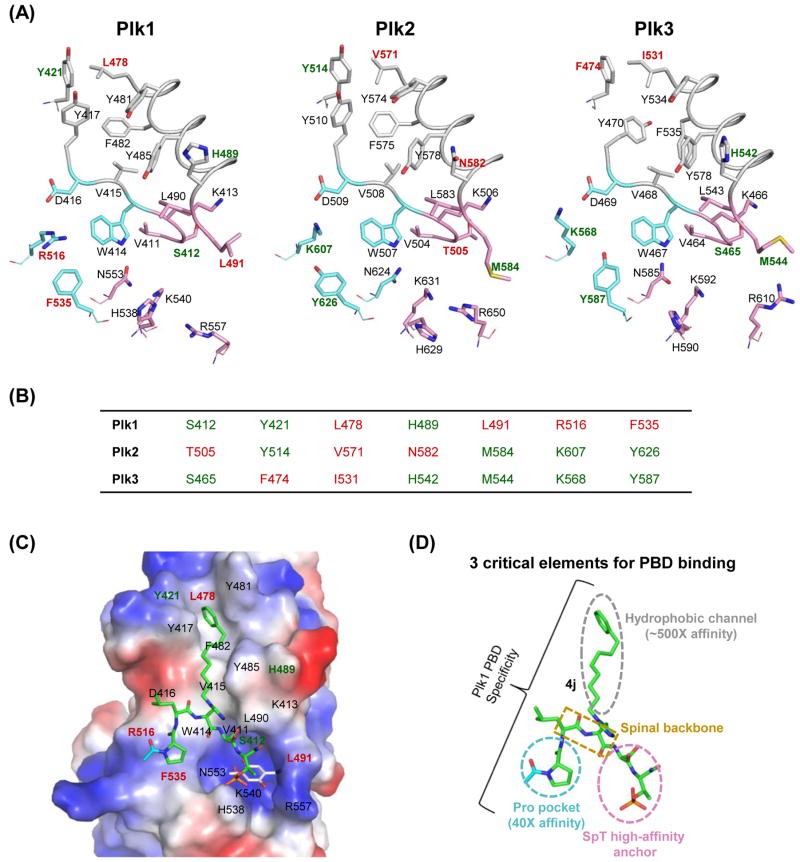
Figure 5.
A structural comparison of Plk1–3 PBDs and the binding mode of Plk1 PBD with its ligands. (A) The apo structures of three Plk1–3 PBDs are represented in stick models: Plk1 PBD (PDB 1UMW), Plk2 PBD (PDB 4RS6), and Plk3 PBD (a model generated using the Robetta server [153]). The PBDs are arbitrarily divided into three regions based on the elements required for 4j binding to Plk1 PBD: a Pro pocket (cyan), a phospho-binding pocket (pink), and a hydrophobic channel (gray). The residues specific to each Plk are shown in red, and the overlapped residues in two of Plk1–3 are indicated in green. (B) Unique residues in each Plk are indicated in red, while residues overlapped in two of Plk1–3 are denoted in green. (C) 4j (green) bound to Plk1 PBD (PDB 3RQ7; [120]) is displayed in an electrostatic surface representation, with overlaid thymoquinone (gray) bound to Plk1 PBD (PDB 4H71; [130]). (D) A schematic diagram showing three elements that are critical for Plk1 PBD binding. The Pro pocket and the hydrophobic channel have been shown to increase the Plk1 PBD-binding affinity ~40-fold and ~500-fold, respectively [82, 120]. The critical role of the SpT anchor has been demonstrated previously [47, 82]. The spinal backbone of 4j is indicated in a dotted rectangular box.