Abstract
Since the 1980s, growing evidence suggested that the cellular localization of proteins determined their activity and biological functions. In a classical view, a protein is characterized by the single cellular compartment where it primarily resides and functions. It is now believed that when proteins appear in different subcellular locations, the cells surpass the expected activity of proteins given the same genomic information to fulfill complex biological behavior. Many proteins are recognized for having the potential to exist in multiple locations in cells. Dysregulation of translocation may cause cancer or contribute to poorer cancer prognosis. Thus, quantitative and comprehensive assessment of dynamic proteins and associated protein movements could be a promising indicator in determining cancer prognosis and efficiency of cancer treatment and therapy. This review will summarize these so-called moonlighting proteins, in terms of a coupled intracellular cancer signaling pathway. Determination of the detailed biological intracellular and extracellular transit and regulatory activity of moonlighting proteins permits a better understanding of cancer and identification of potential means of molecular intervention.
Keywords: Moonlighting protein, translocation, multi-functional proteins, dynamic proteins
1. Introduction
The Human Genome Project determined about 20,000–25,000 human protein-coding genes. Because this number is far lower than predicted numbers of protein (50,000–60,000), the traditional idea of one gene – one protein – one function has become too simple to fully explain the cellular complexity of protein function and interaction. Some partial explanations for this discrepancy are alternative splicing mechanisms, gene fusions, families of homologous proteins, or unrestrained enzyme activities; however, a single protein can have multiple functions that are not result of these phenomena. By providing new insight into the cellular complexity, we can identify a growing number of multifunctional proteins in which the two different functions are found in a single polypeptide. Emerging data on the cellular localization of proteins has exposed additional protein activity that occurs when a protein’s subcellular localization changes from the region where it is first destined. In a classical view, proteins are characterized by a single cellular compartment (cytoplasm, nucleus, plasma membrane, or extracellular region) in which each protein primarily resides and functions [86]. However, unexpected subcellular localization of such proteins challenges this classical view, and gives us one mechanism to explain a protein’s multifunctional ability. For example, the canonical mechanism of extracellular cytokines is to bind to membrane receptors and propagate signals to intracellular effectors; however, these cytokines have also been detected in the nucleus, where they perform an alternative role in transcription. Furthermore, the majority of DNA-binding transcription factors localize in nucleus, but they may also exist in different cellular regions, such as cell membrane and mitochondria, suggesting roles other than as a transcription factor or cofactor. This altered localization may be representative of a new mechanism through which cells can overcome a limited amount of genomic information to fulfill complex biological phenotypes and behavior.
Although many proteins were identified and categorized based on a single activity, each is now known to display multiple, independent functions beyond originally identified ones, and these multifunctional proteins are referred to as “moonlighting” proteins [49]. Moonlighting describes a single protein with multiple functions that are not a result of gene fusions, families of homologous proteins, splice variants, or promiscuous enzyme activities. It is speculated that ancestral moonlighting proteins originally possessed a single function but acquired additional functions through evolution [50]. In addition to a change in cellular location, moonlighting proteins can switch between functions due to a change in temperature, a change in the redox state of the cell, a change in the oligomeric state of the protein, direct interactions with a variety of binding partner proteins, or changes in the cellular concentration of a ligand, substrate, cofactor, or product [44, 51]. Although these switches are all considered as mechanism(s) for moonlighting, this review summarizes well-known proteins where changes in subcellular localization contribute to additional biological activity in cancer.
2. Cytoplasmic proteins (Hsp90, Transglutaminase 2, GAPDH)
2.1. Hsp90
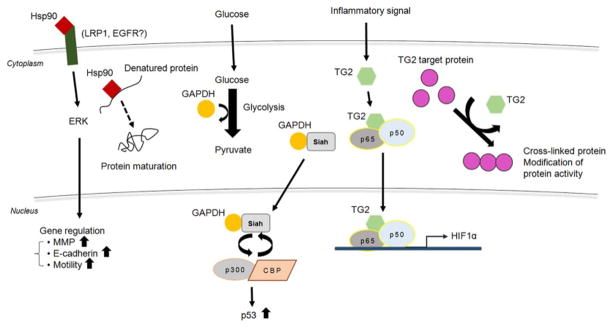
Heat shock protein 90 (Hsp90) is a molecular chaperone, traditionally known cytoplasmic protein that mediates ATP-dependent folding, stabilization, intracellular disposition, and proteolytic turnover of proteins [47, 121]. However, Hsp90 is also a secreted and cell surface protein. Extracellular Hsp90 has long been observed, and its secreted form is considered a pro-tumorigenic protein. Blocking secretion of Hsp90 resulted in a significant inhibition of tumor metastasis, and the serum level of Hsp90 is positively correlated with tumor malignancy in clinical cancer patients [116]. Extracellular Hsp90 interacts with and stabilizes matrix metalloproteinase-2 (MMP-2), which contributes to angiogenesis and cancer cell invasiveness [31, 105]. Secreted Hsp90 has also been reported to regulate and alter E-cadherin function in prostate cancer, giving rise to epithelial-mesenchymal transition (EMT) [40], indicating that it could be a diagnostic marker for tumor malignancy. In addition to this unexpected role of Hsp90 in the extracellular region, Hsp90 also has been found in the nucleus [23] and could regulate several nuclear events [108] (Figure 1). In consideration of such localization, selective Hsp90 inhibitors have been tested for clinical development. These inhibitors bind to the N-terminus of Hsp90 (ATP-binding domain) and induce degradation of multiple oncogenic Hsp90 client proteins, such as HER2/neu [52, 117]. However, further preclinical and clinical studies are required to validate the use of these inhibitors for therapeutic purposes.
Figure 1. Examples of cytosolic moonlighting proteins.
Hsp90, GAPDH, and TG2 are classically known as a cytosolic protein. These proteins also possess biological activity in different translocation such as ECM (Hsp90) and nuclear (GAPDH and TG2). TG2, transglutaminase 2; Hsp90, heat shock protein 90; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Details are in the text.
2.2. Transglutaminase 2
Transglutaminase 2 (TG2), a ubiquitous member of the mammalian transglutaminase family, can catalyze protein crosslinking via transamidation of glutamine residues to lysine residues in a Ca2+-dependent manner [66]. Besides its classical protein crosslinking activity, TG2 possesses several other functions in different cellular compartments, including cell adhesion and kinase activities. TG2 is predominantly a cytoplasmic protein, but increasing evidence indicates that TG2 dynamically translocates depending on cellular context [84], even though the molecular mechanism(s) underlying dynamic translocation of TG2 to various subcellular compartments remains elusive. In addition to transglutaminase (TGase) activity, cytosolic TG2 has been shown to be involved in signal transduction. TG2 activates the NF-κB pathway to elicit anti-apoptotic effects in ovarian cancer cells [11] (Figure 1). TG2 can also function as a G protein (Gh) on the plasma membrane, which has a unique GTP binding pocket and GTP hydrolysis activity, suggesting TG2 may be implicated in receptor signaling [45]. Membrane TG2 have kinase activity and phosphorylate target proteins such as insulin-like growth factor-binding protein-3 (IGFBP-3) [77]. In addition, translocation of TG2 from the cytoplasm to the nucleus was detected in various types of cells [59, 70, 87], implying the versatility of the TG2 protein. Although elevation of intracellular calcium levels promoted TG2 translocation to the nuclear compartment [59], the biological function of nuclear TG2 is still unclear. Emerging evidence indicates the importance of nuclear TG2 in regulating gene expression via post-translational modification of transcription factors (via crosslinking and phosphorylation) and histone proteins. These include E2F1 [82], hypoxia inducible factor 1 [33], Sp1 [106], and all four mammalian core histones (H2A, H2B, H3, and H4) [78]. Although TG2 was originally identified as an intracellular enzyme, TG2 can also be detected in the extracellular space. Extracellular TG2 acts as a matrix stabilizer through crosstalk with other proteins, a component of cell adhesion complex and regulates cell survival [22]. For example, extracellular TG2 is associated with multiple integrins of the β1 and β3 subfamilies and implicated in cell adhesion and spreading [3]. TG2 also interacts with extracellular matrix-associated fibronectin (FN). FN-bound TG2, with increased resilience to MMP degradation, maintains cell adhesion by interacting with cell surface heparin sulfate chains of syndecan-4 [107]. This substantial evidence indicates that various activities of TG2 are differentially regulated depending on TG2’s subcellular localization. Recently, for clinical purposes, large numbers of TG2 inhibitors were screened with respect to adhesion, migration, and invasion of ovarian cancer cells [123].
2.3. GAPDH
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes an important energy-yielding step in glycolysis: GAPDH coverts glyceraldehyde-3-phosphate to D-glycerate 1,3-bisphospate and mediates formation of NADH and adenosine triphosphate (ATP) in the presence of inorganic phosphate and nicotinamide adenine dinucleotide (NAD). In addition to this originally identified function as a “housekeeping” protein in the cytoplasm, emerging data show numerous activities apart from energy production [103, 109]. GAPDH is translocated to the nucleus upon exposure to a stressor, such as oxidative stress, leading to cell death/dysfunction [41]. This nuclear translocation is caused by S-nitrosylation at Cys150 residue and formation of complex with Siah containing the nuclear localization signal (Figure 1). Nuclear GAPDH is further acetylated at Lys160 residue by the acetyltransferase p300/CREB binding protein (CBP) through direct protein interaction, which in turn stimulates acetylation and catalytic activity of p300/CBP, leading to induction of target genes, including tumor suppressor p53 [97]. Various GAPDH nucleic functions that deviate from cell death have been also reported. For example, GAPDH directly interacts with apurinic/apyrimidinic endonuclease 1 (APE1), which is involved in the base excision DNA repair pathway. GAPDH reactivates endonuclease activity of APE1 to cleave a basic sites and to regulate the redox state of a number of transcriptional factors such as p53, AP-1, c-Jun, c-Fos, and NF-κB [5]. GAPDH also forms a complex with OCTamer-binding factor 1 (Oct-1) and p38, and regulate S-phase progression in the cell cycle [24]. Nuclear GAPDH plays a role in maintaining and protecting telomeric DNA from rapid, chemotherapy-induced degradation [26]. Through these findings, GAPDH is not only involved in glycolysis as a cytoplasmic protein to break down glucose for energy and carbon molecules, but also participates in gene regulation as a nuclear factor.
3. Nuclear proteins (HMGB1, p53, ESE-1, and β-catenin)
3.1. HMGB1
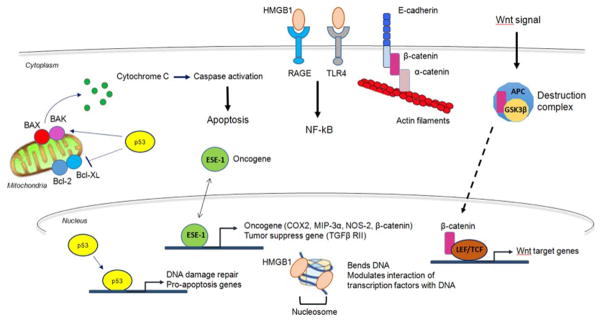
High mobility group box-1 (HMGB1) belongs to one of three families (HMGA, HMGB, and HMGN), and members of each family have been identified as the second-most abundant chromatin proteins participating in gene regulation and cellular differentiation [20]. Moreover, they contribute to the fine tuning of transcription in response to rapid environmental changes by interacting with nucleosomes, transcription factors, nucleosome-remodeling machines, and histone H1 [9]. HMGB1 is a nuclear factor and binds to nucleosome and controls DNA packing and chromatin remodeling. HMGB1 is also a crucial cytokine that mediates the response to infection, injury, and inflammation [67] (Figure 2). HMGB1 is also secreted from cultured macrophages after stimulation with endotoxin, TNF, or IL-1β. Because HMGB1 release occurs considerably later than secretion of the classical early pro-inflammatory mediators TNF and IL-1β, HMGB1 is considered to be a cytokine involved as a late-acting mediator for lethal shock (endotoxemia) [113]. HMGB1 binding to the receptor for advanced glycation end-products (RAGE) and to Toll-like receptors (TLRs) for activating the inflammatory process in immune and endothelial cells [27, 102] has been intensively studied. Secretion of HMGB1 seems not to be restricted to immune cells since epithelial gastric and colon cancer cell lines also show release of HMGB1 [1].
Figure 2. Examples of nuclear moonlighting proteins.
HMGB1, p53, β-catenin, and ESE-1 are classically known as a nuclear protein to control chromatin structure and transcription. These proteins also play significant biological roles in ECM, mitochondria and cytosol. HMGB1, high mobility group box 1. Details are in the text.
3.2. p53 tumor suppressor protein
p53, a well-known tumor suppressor protein, accumulates in the nucleus in response to DNA damage, oncogene activation, and other stress; it acts as a nuclear transcription factor by binding to a specific DNA sequence and regulating a variety of genes [56, 94]. In addition to this nuclear activity, p53 also has different biological activities in the cytoplasmic region–not as a transcription factor [18]. Although mutant p53 loses its transcriptional activity even in the nucleus, it still has the ability to induce apoptosis [126]. In addition, there are reports that p53 rapidly accumulates in the mitochondria under a variety of stress conditions [73]. The mitochondrial membrane is one of the intracellular compartments where cytoplasmic p53 is located under a stress condition, and p53 induces mitochondrial outer membrane permeabilization by triggering the release of pro-apoptotic factors such as Bax and Bak [17, 36]. This indicates that the pro-apoptotic effects of cytoplasmic p53 are independent of transcription. Post-translational modification of p53 and protein interactions can dictate the destination of p53 [34, 73] (Figure 2). These results, taken together, indicate that apoptosis induced by p53 likely arises from combined activities of the cytoplasmic and nuclear proteins. On the other hand, p53 is rapidly degraded by the selective E3 ligase MDM. Therefore, potent and selective small-molecule inhibitors of the p53-MDM2 interaction have been designed with the intention to treat p53 wild-type tumors [68]. Currently, several chemically diverse MDM2-p53 inhibitors are in Phase I or Phase II clinical trials for the treatment of solid and hematological malignancies [127].
3.3. ESE-1
The E26 transformation-specific (ETS) family of transcription factors is composed of more than 40 members and play as a key mediator of morphogenesis and development. Many of them are highly associated with cancer and acts as oncogenes or tumor suppressors. ESE-1 (also known as Elf3, ESX or ERT) is an epithelial specific Ets-1 that is involved in the differentiation of the epithelium. ESE-1 is induced in response to inflammatory cytokines and lipopolysaccharide and enhances transactivation of nitric oxidase synthase 2 (NOS2) synergistically with NF-κB [95] and correlate to induction of cyclooxygenase-2 (COX-2) [35] by pro-inflammatory cytokines and leads to inflammatory bowel disease. ESE-1 also directly activates expression of TGF-β receptor type II (TGF-β RII) and inhibits TGF-β resistance [19, 85]. Recently, there has been a focus on the dichotomous role of ESE-1 in tumorigenesis. Many studies claimed that overexpression of ESE-1 induced cell proliferation, adhesion, invasion and epithelial to mesenchymal transition (EMT). Stable expression of ESE-1 leads to transformation of breast cancer cells and a knockdown of ESE-1 decreased cancer cell growth [112]. In addition, expression of ESE-1 is associated with poor prognosis of colon [114] and prostate cancer [65]. In terms of oncogenic mechanism, ESE-1 is a target of oncogenic kinase, PAK1 and phosphorylated ESE-1 increase stabilize and cause transformation of breast cancer cells [69]. On the contrary, increased expression of ESE-1 inhibited tumorigenesis in prostate cancer cells [99] and resulted in reduced tumorigenicity through the TGF-β-mediated pathway [14]. In terms of two distinct ESE-1 function, Prescott et al., reported that cytosolic ESE-1 leads to transformation of breast cancer through SAR domain-mediated pathway, whereas nuclear ESE-1 increase apoptosis through transcriptional-mediated pathway [90]. Recently, we reported that treatment of anti-cancer compounds such as tolfenamic acid leads to nuclear translocation of ESE-1 [58], which might be a potential mechanism of apoptosis. In cancer cells, ESE-1 co-localizes both in the cytosol and in the nucleus, and it may dynamically translocate depending on cellular context although the molecular mechanism of translocation remains obscure. Further studies will need to clarify the biological function of ESE-1 depending on cellular localization
3.4. β-catenin
Wnt signaling is essential for embryogenesis, tissue development, cell proliferation and differentiation. β-catenin is a key mediator of Wnt signaling and Wnt-specific gene expression that controls cell fate decisions. The nuclear function of β-catenin as a transcription co-factor has been well studied in colon cancer development, and abnormal Wnt signaling has been linked to adenomatous polyposis coli (APC) mutations. These mutations lead to an increase in cell proliferation and migration, stem cell-self renewal, and EMT events [93, 96, 104]. Therefore, truncated germline mutations of APC are very frequent in patients with familial adenomatous polyposis and progress colorectal carcinomas in combination with mutations in KRAS and TP53 [104]. Nuclear localization of β-catenin and transcription activity is tightly regulated by Wnt signaling and proteosomal degradation of β-catenin. In the absence of Wnt ligands, cytoplasmic β-catenin is subjected to form a destruction complex composed of various proteins and is phosphorylated by GSK3β and CK1. As a consequence, β-catenin is ubiquitinated and targeted for degradation by proteasomes. Alternatively, upon Wnt receptor activation, the destruction complex is recruited to the Wnt receptor, which causes GSK3β and CK1 inactivation. As a result, β-catenin phosphorylation is inhibited, and stabilized surplus β-catenin escaping ubiquitination translocates into the nucleus, where it engages with lymphoid enhancer factor/T-cell factor (TCF) transcription factors to control Wnt-specific gene expression. Thus, mutations in a destruction complex component, such as APC, could facilitate the stabilization and accumulation of β-catenin in the nucleus, which contributes to Wnt signal activation and carcinogenesis [88]. If β-catenin forms a complex together with E-cadherin at the plasma membrane, this complex could mediate the interplay of junction molecules with the actin cytoskeleton [111]. Changes in cadherin-based adherens junctions contribute to the disruption of epithelial cell polarity, which is the onset of morphology change processes such as tissue rearrangement, cell migration and differentiation, EMT, and metastasis [74]. Recently, novel chemical inhibitors of β-catenin were identified. For example, CWP232228 antagonizes binding of β-catenin to TCF in the nucleus and suppresses breast cancer stem-like cells, implying potential use of these small molecular inhibitors as a cancer therapy [48].
4. Secreted proteins (MMPs and NAG-1)
4.1. MMPs
Matrix metalloproteinases (MMPs) were originally identified as zinc-dependent endopeptidases that act in the extracellular matrix. Most studies on the biological function of MMPs focused on degradation and turnover of extracellular matrix (ECM) components which contribute to cell invasion [55, 83]. Emerging data have uncovered nontraditional roles for MMPs in the extracellular space as well as in the nucleus [71]. The biological role of intracellular MMPs and their mechanism(s) for protein trafficking are still unclear, although recent studies have demonstrated some of their functions in intracellular regions and mechanism(s) for protein trafficking. Specifically, the presence of MMP-2 in the nucleus of cardiac myocytes has been reported. Poly (ADP-ribose) polymerase (PARP) may be a nuclear substrate of MMP-2, suggesting a possible role of nuclear MMP-2 in PARP degradation [57]. MMP-3 is also found in the nucleus in several cultured cell types and in human liver tissue sections. MMP-3 may have a putative NLS at position 107 to 113 in the amino acid sequence which is responsible for nucleus entry through the nuclear pore via a mechanism involving transporter protein recognition of NLS. Nuclear expression of MMP3 is associated with an increased rate of apoptosis, and MMP3 appears to serve as an anti-tumorigenic protein [101]. An additional putative NLS in MMP3 and unique function of nuclear MMP3 as a transcriptional factor has been identified (Figure 3). MMP-3 binds to a enhancer of the connective tissue growth factor (CCN2/CTGF) promoter, transactivates the CCN2/CTGF gene [28], and promotes chondrocyte proliferation and ECM remodeling. These studies suggest a new role of MMP3 in the development, tissue remodeling, and pathology of arthritic diseases through CCN2/CTGF regulation. MMP-12 is known as a macrophage metalloelastase which contributes to degradation of the extracellular matrix during inflammatory tissue destruction. However, MMP-12 can be translocated into the nucleus where it binds to the NFKBIA promoter and drive transcription [72]. Intracellular MMP-12 mediates NFKBIA transcription, leading to IFN-α secretion and host protection from virus infection. The recent findings of nuclear localization of MMPs open a new avenue of study in which MMPs cleave and activate intracellular peptides as well as induce gene expression.
Figure 3. Examples of secreted moonlighting proteins.
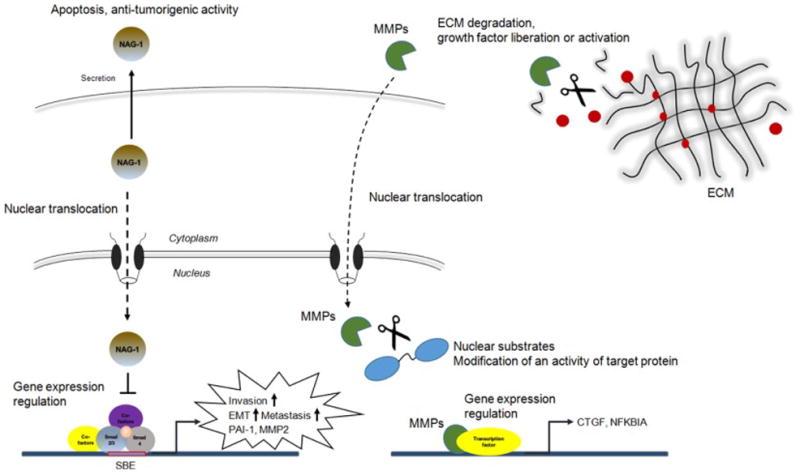
NAG-1 and MMPs are classically known as a secreted proteins. However, these proteins also perform biological activity in different translocation. NAG-1, Nonsteroindal anti-inflammatory drug activated gene-1; MMPs, Matrix metalloproteinases.
4.2. NAG-1
We identified the NSAID-activated gene-1 (NAG-1, also known as GDF15) as a divergent member of the TGF-β superfamily by PCR-based subtractive hybridization from an NSAID-induced library in cyclooxygenase (COX)-negative cells [6]. NAG-1 expression reduces TNF-α secretion in macrophages [10], and ectopic expression of NAG-1 causes cell growth arrest [6, 8]. Furthermore, overexpression of NAG-1 in human colon, bladder, and glioblastoma cells inhibits tumor formation in the nude mouse model [4, 6, 110]. The principal function, receptor, and signaling pathway of NAG-1 remain uncertain in cancer, and the biological role of NAG-1 in diseases remains poorly understood with sometimes contradictory evidence. Several studies have observed major up-regulation of NAG-1 mRNA and protein in cancer biopsies [81, 98], whereas a number of other studies, including ours, have demonstrated an anti-tumorigenic function for NAG-1, in which NAG-1 induces apoptosis and negatively affects tumor growth [6, 7, 16, 53, 54, 124]. Although in vitro assays show different results, studies conducted in NAG-Tg and NAG-1 KO mice consistently demonstrate anti-tumorigenic activity associated with expression of this protein [7, 12, 128]. Some possible explanations of the contradictory activity in vitro include 1) NAG-1’s different functions in different cancer types, 2) an unidentified role of pro-mature (pre-processed) NAG-1 in cells, and 3) the contribution of NAG-1 binding proteins or receptors in different cells. Overall, NAG-1 represents a highly promising molecular target for anti-cancer activity induced by NSAIDs, and elucidation of its cellular movement still needs to be determined toward this end. NAG-1 is synthesized as 308-amino acid pro-NAG-1 and then dimerized by specific disulfide linkage. A pro-NAG-1 dimer is then cleaved by furan-like proteases at an RXXR site, forming a 112 amino acid C-terminal dimeric protein and a pro-peptide [115]. This mature dimeric protein is secreted into the extracellular matrix and can be found in human blood. Experimental evidence clearly confirms the secreted mature dimer has biological activity [21]; although pro-NAG-1 is present in cells, its fate and role resulting in biological activity is unclear. We have recently reported that pro-NAG-1 accumulates in the nucleus and contributes to transcriptional regulation [76]. Pro-NAG-1 is expressed in not only the cytoplasm and ECM, but also in the nucleus. NAG-1 is dynamically moved to the nucleus, exported into cytoplasm, and further transported into the ECM (Figure 3). We have also found that nuclear NAG-1 contributes to inhibition of the Smad pathway by interrupting the binding of the Smad complex to DNA.
5. Other proteins (E-cadherin, TGFα receptor I, EGFR, and Smad)
5.1. E-Cadherin
E-cadherin is a class I transmembrane glycoprotein that plays important roles in cell adhesion and formation of adherens junctions. The classically and originally identified function of this protein was as a cell membrane protein [37, 38]. E-cadherin consists of 5 cadherin repeats (EC1 – EC5) in the extracellular domain that bind calcium ions to form a stiffened linear molecule, one transmembrane domain, and an intracellular domain that interacts with catenins and a variety of actin-binding proteins to anchor the cadherin–catenin complex to the actin cytoskeleton [37]. E-cadherin can also block growth factor-mediated proliferation signaling (contact inhibition of growth), thereby maintaining tissue integrity and preserving tissue function [91]. A decrease in E-cadherin, attenuating strong cell-cell interactions, has been implicated in cancer progression and metastasis [89]. Decreasing the strength of cellular adhesion to a tissue gives rise to cancer cell motility, enabling cancer cells to cross the basement membrane and invade surrounding tissues. Reduced expression of E-cadherin is indicative of an unfavorable clinical outcome in several malignant diseases [125]. This evidence supports the notion that E-cadherin may be a tumor-suppressor protein. However, emerging roles of nuclear E-cadherin as a modulator of tumor growth survival and motility, triggering cancer progression, has been reported; therefore, E-cadherin may also possess oncogenic functions. E-cadherin has been shown to be cleaved by γ-secretase, after which the cytoplasmic fragment of E-cadherin translocates to the nucleus and regulates gene expression [32], implying that oncogenic functions of E-cadherin may arise from the cleaved form in the nucleus. The nuclear E-cadherin fragment interacts with CTF2 to suppress the induction of apoptosis [32]. In vivo study has shown that detection of aberrant nuclear E-cadherin correlates with lymph node spread and liver metastases in pancreatic endocrine tumors [15]. In addition to γ-secretase, several proteases, and matrix metalloproteinases (MMP-3, MMP-7, MMP-9, and MT1-MMP) have also been reported to convert the mature 120 kDa E-cadherin into an extracellular N-terminal 80-kDa fragment and an intracellular C-terminal 38-kDa fragment [25]. Interestingly, the extracellular fragment is released from the plasma membrane and diffuses into the extracellular environment and even the bloodstream to serve as a paracrine/autocrine signaling molecule [46].
5.2. TGF-β receptor I
TGFβ receptors are type I transmembrane proteins that possess serine/threonine kinase activity. In the canonical pathway, active TGF-β1 binds to cell surface receptor kinases TGF-β type I (TβRI) and type II receptors (TβRII). Upon ligand binding to TβRII, TβRII further phosphorylates and activates TβRI, which in turn propagates the signals by phosphorylating the aforementioned Smad proteins [100] to regulate cell differentiation, morphogenesis, tissue homeostasis, and regeneration. This function as a cell membrane receptor for sensing and propagating signals was originally identified as the role of TGFβ receptors. Since TNF-alpha converting enzyme (TACE) has recently been shown to cleave and release ectodomains of TβRI and attenuate TGFβ signaling [62], Yabing Mu’s group reported that TGFβ induces Lys63-linked polyubiquitination of TβRI via TRAF6 to promote cleavage of TβRI by the metalloprotease TACE, resulting in translocation of the intracellular domain of TβRI to the nucleus [79]. Once the liberated intracellular domain (ICD) of TβRI is translocated to the nucleus, it associates with the transcriptional regulator p300 and promotes tumor progression by activating invasion-regulating target genes such as Snail and MMP2. Nuclear translocation of TβRI in response to TGFβ is not abolished by TβRI serine/threonine kinase inhibitor SB505124, implying that a therapeutic strategy that targets both a kinase function and nuclear function of TβRI may elicit a better effect in inhibiting tumor progression. Another recent report has shown that ligand-stimulated TβRI is translocated to the nucleus in association with importin β1, nucleolin, and Smad2/3 in HER2-transformed cells. In the nucleus, TβRI specifically recognizes RNA targets and regulates RNA processing [13].
5.3. EGFR
The epidermal growth factor receptor (EGFR, also known as ErbB1/HER1) is a member of the EGFR tyrosine kinase family and consists of HER2/neu (ErbB2), HER3 (ErbB3), and HER4 (ErbB4). EGFR triggers intracellular signaling pathways by phosphorylating downstream signal molecules such as the RAS/MAPK, PI(3)K/Akt, PLCγ/PKC, and Jak/STAT. Stimulation of these kinases and downstream cell signaling cascades influence cell proliferation, apoptosis, migration, and survival, as well as complex biological processes. There are many reports that EGFR has been consistently detected in the nuclei of cancer cells from primary tumor specimens and highly proliferative tissues [61, 122], suggesting a second function of EGFR as a nuclear factor in addition to its role in traditional cytoplasmic signaling. Research over the last decade has demonstrated the mechanism(s) of transport of EGFR to the nucleus. The EGFR family members (EGFR, ErbB2, ErbB3, and ErbB4) have a conserved tripartite nuclear localization signal (NLS), which can recognize importin β1 for shuttling EGFR to the nucleus [43]. EGFR is described as undergoing COPI-mediated retrograde trafficking from the Golgi apparatus to the ER [119], and then EGFR is translocated to the inner nuclear membrane (INM). Molecular mediators for nuclear translocation of EGFR include dynamin, importins, and Sec61 [118], and CRM1 exportin may be involved in the nuclear export of EGFR [63]. Upon entry into the nucleus, EGFR can function in ways distinct from the receptor tyrosine kinase on plasma membrane. Many reports have indicated a role of nuclear EGFR as a co-transcription factor via its interaction with transcription factors, including E2F1 and STAT3 [29, 39, 64]. This evidence suggests that the subcellular distribution of EGFR contributes to the evocation of different biological outcomes that need to be considered in the field of EGFR studies. Indeed, a recent study demonstrated that cetuximab, which is an IgG1 chimeric monoclonal antibody targeting the ligand-binding domain of the EGFR, may have varied efficacy due to the presence of nuclear EGFR. Although cetuximab can abrogate signals from plasma membrane EGFR, nuclear EGFR is not affected by cetuximab. Therefore, nuclear EGFR is not targeted by cetuximab and propagates its own signals through the modulation of cyclin D1, B-myb, and Aurora kinase A, contributing to increased proliferation [60]. This implies that new therapeutic chemicals need to be considered to target both plasma membrane EGFR and nuclear EGFR.
5.4. Smads
Smad family transcription factors are downstream mediators of TGFβ superfamily cytokines, and eight Smad proteins have been identified in human and mouse [75]. Smad1, Smad2, Smad3, Smad5, and Smad8 are substrates for TGFβ receptors, referred to as receptor-regulated Smads, or R-Smads. Smad4 interacts with all R-Smads as a common partner, also referred to as Co-Smad. Smad6 and Smad7 are inhibitory Smads, also referred to as I-Smads that attenuate interaction between Smad-receptors or Smad-Smad interaction [2]. R-Smads constantly shuttle between the cytoplasm and the nucleus in the basal state to monitor if TGFβ receptors are activated. Upon ligand binding, R-Smad phosphorylated by its receptor can be recognized by the Co-Smad (Smad4); in this cellular context, the complex then enters the nucleus more rapidly to regulate transcription of target gene incorporation with other DNA-binding cofactors [42, 80]. This process originally characterized R-Smads and Smad4 in the nucleus as playing a role in transcription factor-mediated TGFβ signaling. However, Smad could be an adaptor protein in the cytoplasm to regulate other signal pathways. For example, Dvl-1, Erbin, and Par-3 have been identified as Smad3 binding proteins using a yeast two-hybrid screen, suggesting that Smads can directly interact with these proteins not as transcription factors, but to regulate cell polarity [120]. Smad3 also binds to collagen types I, III, and V, implying another aspect of Smad proteins not serving as a transcription factor in the development of the orofacial region [30]. Smad4 binds to Hoxa9 protein in the cytoplasm to prevent nuclear translocation of Hoxa9, suggesting a protective role of Smad4 against nuclear activation by Hoxa9 and leukemia transformation [92]. These examples indicate that monitoring the activation of the TGFβ receptor is not only a task that Smad proteins have for ultimate regulation of gene expression, but Smad proteins also interact with different components of other pathways in the cytoplasm [129] as adaptor or anchor proteins for fine tuning and crosstalk of the TGFβ signal with anther signaling pathway.
6. Conclusion
The list of multifunctional proteins and their new biological roles continue to grow. Changing subcellular localization contributes to giving a protein additional function, as seen in the proteins described above. The take-home message of this review is that disease progress may not only arise from a mutation in a gene and expression of splice variants, but also from dysregulation of the protein sorting system that controls protein localization in the cell. These findings articulate a need for further study of the molecular mechanisms by which moonlighting proteins affect biologic activity through protein production, modification, secretion, and nuclear translocation to interact with other proteins in the cell.
Table 1.
Examples of multifunctional proteins shown in cancer and their subcellular localizations and functions.
| Protein | Subcellular localization/function
|
|
|---|---|---|
| Primary | Additional | |
| p53 | Nucleus/Transcription factor | Mitochondria/Promoting mitochondrial membrane permeabilization |
| Smads | Nucleus/Transcription factor | Cytoplasm/Adaptor protein |
| Hsp90 | Cytoplasm/Molecular chaperone | Extracellular region/Regulator for angiogenesis and cell invasiveness |
| TG2 | Cytoplasm/Catalyzes protein cross-linking | (1) Nucleus/Regulates gene expression; (2) Extracellular region/Cell adhesion and spreading |
| β-catenin | Nucleus/Transcription factor | Cytoplasm/Interaction with catenins and actins |
| GAPDH | Cytoplasm/Glycolytic enzyme | Nucleus/Participates in gene regulation |
| HMGB1 | Nucleus/Binds to nucleosomes | Extracellular region/Inflammatory cytokine |
| E-cadherin | Cell surface/Involved in cell adhesion | Nucleus/Gene regulation |
| TβRI | Cell surface/Type I transmembrane protein | Nucleus/Participates in gene regulation |
| EGFR | Cell surface/Membrane receptor tyrosine kinase for EGF | Nucleus/Co-transcription factor |
| MMPs | Extracellular region/Secretion protein involved in proteolysis | Nucleus/Regulates gene expression |
| NAG-1 | Extracellular region/Secretion protein involved in apoptosis | Nucleus/Gene regulation |
| ESE-1 | Nucleus/Morphology and terminal differentiation | Cytosol/Transformation |
Highlights.
We summarized moonlighting proteins in cancer signaling.
Classical localization of cytoplasmic proteins, nuclear proteins, secreted proteins, membrane bound proteins, and receptors were described with alterative localization.
Acknowledgments
We apologize to all colleagues whose important work we could not cite due to space restrictions. We thank Ms. Misty Bailey (University of Tennessee) for her critical reading of this manuscript. This work was supported by The University of Tennessee Center of Excellence in Livestock Diseases and Human Health grant and NIH grant R01CA108975 (SJ Baek).
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abe A, Kuwata T, Yamauchi C, Higuchi Y, Ochiai A. High Mobility Group Box1 (HMGB1) released from cancer cells induces the expression of pro-inflammatory cytokines in peritoneal fibroblasts. Pathol Int. 2014;64:267–275. doi: 10.1111/pin.12167. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst RJ, Hata A. Targeting the TGF[beta] signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akimov SS, Belkin AM. Cell surface tissue transglutaminase is involved in adhesion and migration of monocytic cells on fibronectin. Blood. 2001;98:1567–1576. doi: 10.1182/blood.v98.5.1567. [DOI] [PubMed] [Google Scholar]
- 4.Albertoni M, Shaw PH, Nozaki M, Godard S, Tenan M, Hamou MF, Fairlie DW, Breit SN, Paralkar VM, de Tribolet N, Van Meir EG, Hegi ME. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21:4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- 5.Azam S, Jouvet N, Jilani A, Vongsamphanh R, Yang X, Yang S, Ramotar D. Human Glyceraldehyde-3-phosphate Dehydrogenase Plays a Direct Role in Reactivating Oxidized Forms of the DNA Repair Enzyme APE1. J Biol Chem. 2008;283:30632–30641. doi: 10.1074/jbc.M801401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek SJ, Kim KS, Nixon JB, Wilson LC, Eling TE. Cyclooxygenase inhibitors regulate the expression of a TGF-beta superfamily member that has proapoptotic and antitumorigenic activities. Mol Pharmacol. 2001;59:901–908. [PubMed] [Google Scholar]
- 7.Baek SJ, Okazaki R, Lee SH, Martinez J, Kim JS, Yamaguchi K, Mishina Y, Martin DW, Shoieb A, McEntee MF, Eling TE. Nonsteroidal anti-inflammatory drug-activated gene-1 over expression in transgenic mice suppresses intestinal neoplasia. Gastroenterology. 2006;131:1553–1560. doi: 10.1053/j.gastro.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Baek SJ, Wilson LC, Eling TE. Resveratrol enhances the expression of non-steroidal anti-inflammatory drug-activated gene (NAG-1) by increasing the expression of p53. Carcinogenesis. 2002;23:425–434. doi: 10.1093/carcin/23.3.425. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY, Zhang HP, Donnellan M, Mahler S, Pryor K, Walsh BJ, Nicholson RC, Fairlie WD, Por SB, Robbins JM, Breit SN. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci U S A. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao L, Petrusca DN, Satpathy M, Nakshatri H, Petrache I, Matei D. Tissue transglutaminase protects epithelial ovarian cancer cells from cisplatin-induced apoptosis by promoting cell survival signaling. Carcinogenesis. 2008;29:1893–1900. doi: 10.1093/carcin/bgn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cekanova M, Lee SH, Donnell RL, Sukhthankar M, Eling TE, Fischer SM, Baek SJ. Nonsteroidal anti-inflammatory drug-activated gene-1 expression inhibits urethane-induced pulmonary tumorigenesis in transgenic mice. Cancer Prev Res (Phila Pa) 2009;2:450–458. doi: 10.1158/1940-6207.CAPR-09-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra M, Zang S, Li H, Zimmerman LJ, Champer J, Tsuyada A, Chow A, Zhou W, Yu Y, Gao H, Ren X, Lin RJ, Wang SE. Nuclear Translocation of Type I Transforming Growth Factor β Receptor Confers a Novel Function in RNA Processing. Mol Cell Biol. 2012;32:2183–2195. doi: 10.1128/MCB.00320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J, Lee C, Hahm KB, Yi Y, Choi SG, Kim SJ. Over-expression of ERT(ESX/ESE-1/ELF3), an ets-related transcription factor, induces endogenous TGF-beta type II receptor expression and restores the TGF-beta signaling pathway in Hs578t human breast cancer cells. Oncogene. 2000;19:151–154. doi: 10.1038/sj.onc.1203252. [DOI] [PubMed] [Google Scholar]
- 15.Chetty R, Serra S, Asa SL. Loss of Membrane Localization and Aberrant Nuclear E-cadherin Expression Correlates With Invasion in Pancreatic Endocrine Tumors. The Am J Surg Pathol. 2008;32:413–419. doi: 10.1097/PAS.0b013e31813547f8. [DOI] [PubMed] [Google Scholar]
- 16.Chintharlapalli S, Papineni S, Baek SJ, Liu S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes are peroxisome proliferator-activated receptor gamma agonists but decrease HCT-116 colon cancer cell survival through receptor-independent activation of early growth response-1 and nonsteroidal anti-inflammatory drug-activated gene-1. Mol Pharmacol. 2005;68:1782–1792. doi: 10.1124/mol.105.017046. [DOI] [PubMed] [Google Scholar]
- 17.Chipuk JE. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 18.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 19.Choi SG, Yi Y, Kim YS, Kato M, Chang J, Chung HW, Hahm KB, Yang HK, Rhee HH, Bang YJ, Kim SJ. A novel ets-related transcription factor, ERT/ESX/ESE-1, regulates expression of the transforming growth factor-beta type II receptor. J Biol Chem. 1998;273:110–117. doi: 10.1074/jbc.273.1.110. [DOI] [PubMed] [Google Scholar]
- 20.Chou DKH, Evans JE, Jungalwala FB. Identity of nuclear high-mobility-group protein, HMG-1, and sulfoglucuronyl carbohydrate-binding protein, SBP-1, in brain. J Neurochem. 2001;77:120–131. doi: 10.1046/j.1471-4159.2001.t01-1-00209.x. [DOI] [PubMed] [Google Scholar]
- 21.Chrysovergis K, Wang X, Kosak J, Lee SH, Kim JS, Foley JF, Travlos G, Singh S, Baek SJ, Eling TE. NAG-1/GDF-15 prevents obesity by increasing thermogenesis, lipolysis and oxidative metabolism. Int J Obes (Lond) 2014;38:1555–1564. doi: 10.1038/ijo.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collighan RJ, Griffin M. Transglutaminase 2 cross-linking of matrix proteins: biological significance and medical applications. Amino Acids. 2009;36:659–670. doi: 10.1007/s00726-008-0190-y. [DOI] [PubMed] [Google Scholar]
- 23.Csermely P, Schnaider T, Sőti C, Prohászka Z, Nardai G. The 90-kDa Molecular Chaperone Family: Structure, Function, and Clinical Applications. A Comprehensive Review. Pharmacol Therapeut. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 24.Dai RP, Yu FX, Goh SR, Chng HW, Tan YL, Fu JL, Zheng L, Luo Y. Histone 2B (H2B) Expression Is Confined to a Proper NAD+/NADH Redox Status. J Biol Chem. 2008;283:26894–26901. doi: 10.1074/jbc.M804307200. [DOI] [PubMed] [Google Scholar]
- 25.David JM, Rajasekaran AK. Dishonorable Discharge: The Oncogenic Roles of Cleaved E-Cadherin Fragments. Cancer Res. 2012;72:2917–2923. doi: 10.1158/0008-5472.CAN-11-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demarse NA, Ponnusamy S, Spicer EK, Apohan E, Baatz JE, Ogretmen B, Davies C. Direct Binding of Glyceraldehyde 3-Phosphate Dehydrogenase to Telomeric DNA Protects Telomeres against Chemotherapy-Induced Rapid Degradation. J Mol Biol. 2009;394:789–803. doi: 10.1016/j.jmb.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diener KR, Al-Dasooqi N, Lousberg EL, Hayball JD. The multifunctional alarmin HMGB1 with roles in the pathophysiology of sepsis and cancer. Immunol Cell Biol. 2013;91:443–450. doi: 10.1038/icb.2013.25. [DOI] [PubMed] [Google Scholar]
- 28.Eguchi T, Kubota S, Kawata K, Mukudai Y, Uehara J, Ohgawara T, Ibaragi S, Sasaki A, Kuboki T, Takigawa M. Novel transcription factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol Cell Biol. 2008;28:2391–2413. doi: 10.1128/MCB.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eldredge ER, Korf GM, Christensen TA, Connolly DC, Getz MJ, Maihle NJ. Activation of c-fos gene expression by a kinase-deficient epidermal growth factor receptor. Mol Cell Biol. 1994;14:7527–7534. doi: 10.1128/mcb.14.11.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis LR, Warner DR, Greene RM, Pisano MM. Interaction of Smads with collagen types I, III, and V. Biochem Bioph Res Comm. 2003;310:1117–1123. doi: 10.1016/j.bbrc.2003.09.130. [DOI] [PubMed] [Google Scholar]
- 31.Eustace BK, Sakurai T, Stewart JK, Yimlamai D, Unger C, Zehetmeier C, Lain B, Torella C, Henning SW, Beste G, Scroggins BT, Neckers L, Ilag LL, Jay DG. Functional proteomic screens reveal an essential extracellular role for hsp90[alpha] in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 32.Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, Daniel J, Fujita Y. A Role for the Cleaved Cytoplasmic Domain of E-cadherin in the Nucleus. J Biol Chem. 2008;283:12691–12700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filiano AJ, Bailey CDC, Tucholski J, Gundemir S, Johnson GVW. Transglutaminase 2 protects against ischemic insult, interacts with HIF1β, and attenuates HIF1 signaling. FASEB J. 2008;22:2662–2675. doi: 10.1096/fj.07-097709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foo RS. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci USA. 2007;104:20826–20831. doi: 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grall FT, Prall WC, Wei W, Gu X, Cho JY, Choy BK, Zerbini LF, Inan MS, Goldring SR, Gravallese EM, Goldring MB, Oettgen P, Libermann TA. The Ets transcription factor ESE-1 mediates induction of the COX-2 gene by LPS in monocytes. FEBS J. 2005;272:1676–1687. doi: 10.1111/j.1742-4658.2005.04592.x. [DOI] [PubMed] [Google Scholar]
- 36.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 38.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Gene Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 39.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 40.Hance MW, Dole K, Gopal U, Bohonowych JE, Jezierska-Drutel A, Neumann CA, Liu H, Garraway IP, Isaacs JS. Secreted Hsp90 Is a Novel Regulator of the Epithelial to Mesenchymal Transition (EMT) in Prostate Cancer. J Biol Chem. 2012;287:37732–37744. doi: 10.1074/jbc.M112.389015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 42.Hill CS. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 2009;19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 43.Hsu SC, Hung MC. Characterization of a Novel Tripartite Nuclear Localization Sequence in the EGFR Family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 44.Huberts DHEW, van der Klei IJ. Moonlighting proteins: An intriguing mode of multitasking. BBA - Mol Cell Res. 2010;1803:520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 45.Iismaa SE, Wu MJ, Nanda N, Church WB, Graham RM. GTP Binding and Signaling by Gh/Transglutaminase II Involves Distinct Residues in a Unique GTP-binding Pocket. J Biol Chem. 2000;275:18259–18265. doi: 10.1074/jbc.M000583200. [DOI] [PubMed] [Google Scholar]
- 46.Inge LJ, Barwe SP, D’Ambrosio J, Gopal J, Lu K, Ryazantsev S, Rajasekaran SA, Rajasekaran AK. Soluble E-cadherin promotes cell survival by activating epidermal growth factor receptor. Exp Cell Res. 2011;317:838–848. doi: 10.1016/j.yexcr.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 47.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 48.Jang GB, Hong IS, Kim RJ, Lee SY, Park SJ, Lee ES, Park JH, Yun CH, Chung JU, Lee KJ, Lee HY, Nam JS. Wnt/β-Catenin Small-Molecule Inhibitor CWP232228 Preferentially Inhibits the Growth of Breast Cancer Stem-like Cells. Cancer Res. 2015;75:1691–1702. doi: 10.1158/0008-5472.CAN-14-2041. [DOI] [PubMed] [Google Scholar]
- 49.Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 50.Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19:415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- 51.Jeffery CJ. Moonlighting proteins-an update. Mol BioSyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 52.Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. BBA - Mol Cell Res. 2012;1823:742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jutooru I, Chadalapaka G, Chintharlapalli S, Papineni S, Safe S. Induction of apoptosis and nonsteroidal anti-inflammatory drug-activated gene 1 in pancreatic cancer cells by a glycyrrhetinic acid derivative. Mol Carcinog. 2009;48:692–702. doi: 10.1002/mc.20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly JA, Scott Lucia M, Lambert JR. p53 controls prostate-derived factor/macrophage inhibitory cytokine/NSAID-activated gene expression in response to cell density, DNA damage and hypoxia through diverse mechanisms. Cancer Lett. 2009;277:38–47. doi: 10.1016/j.canlet.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 55.Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruse J-P, Gu W. Modes of p53 Regulation. Cell. 137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwan JA, Schulze CJ, Wang W, Leon H, Sariahmetoglu M, Sung M, Sawicka J, Sims DE, Sawicki G, Schulz R. Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of cardiac myocytes and is capable of cleaving poly (ADP-ribose) polymerase (PARP) in vitro. FASEB J. 2004;18:690–692. doi: 10.1096/fj.02-1202fje. [DOI] [PubMed] [Google Scholar]
- 58.Lee SH, Bahn JH, Choi CK, Whitlock NC, English AE, Safe S, Baek SJ. ESE-1/EGR-1 pathway plays a role in tolfenamic acid-induced apoptosis in colorectal cancer cells. Mol Cancer Ther. 2008;7:3739–3750. doi: 10.1158/1535-7163.MCT-08-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lesort M, Attanavanich K, Zhang J, Johnson GVW. Distinct Nuclear Localization and Activity of Tissue Transglutaminase. J Biol Chem. 1998;273:11991–11994. doi: 10.1074/jbc.273.20.11991. [DOI] [PubMed] [Google Scholar]
- 60.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 62.Liu C, Xu P, Lamouille S, Xu J, Derynck R. TACE-Mediated Ectodomain Shedding of the Type I TGF-β Receptor Downregulates TGF-β Signaling. Mol cell. 2009;35:26–36. doi: 10.1016/j.molcel.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin β1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 64.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Longoni N, Sarti M, Albino D, Civenni G, Malek A, Ortelli E, Pinton S, Mello-Grand M, Ostano P, D’Ambrosio G, Sessa F, Garcia-Escudero R, Thalmann GN, Chiorino G, Catapano CV, Carbone GM. ETS transcription factor ESE1/ELF3 orchestrates a positive feedback loop that constitutively activates NF-kappaB and drives prostate cancer progression. Cancer Res. 2013;73:4533–4547. doi: 10.1158/0008-5472.CAN-12-4537. [DOI] [PubMed] [Google Scholar]
- 66.Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 67.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 68.Lv PC, Sun J, Zhu HL. Recent advances of p53-MDM2 small molecule inhibitors (2011-present) Curr Med Chem. 2015;22:618–626. doi: 10.2174/0929867322666141128162557. [DOI] [PubMed] [Google Scholar]
- 69.Manavathi B, Rayala SK, Kumar R. Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. J Biol Chem. 2007;282:19820–19830. doi: 10.1074/jbc.M702309200. [DOI] [PubMed] [Google Scholar]
- 70.Mann AP, Verma A, Sethi G, Manavathi B, Wang H, Fok JY, Kunnumakkara AB, Kumar R, Aggarwal BB, Mehta K. Overexpression of Tissue Transglutaminase Leads to Constitutive Activation of Nuclear Factor-κB in Cancer Cells: Delineation of a Novel Pathway. Cancer Res. 2006;66:8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 71.Mannello F, Medda V. Nuclear localization of Matrix metalloproteinases. Prog Histochem Cytochem. 2012;47:27–58. doi: 10.1016/j.proghi.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Marchant DJ, Bellac CL, Moraes TJ, Wadsworth SJ, Dufour A, Butler GS, Bilawchuk LM, Hendry RG, Robertson AG, Cheung CT, Ng J, Ang L, Luo Z, Heilbron K, Norris MJ, Duan W, Bucyk T, Karpov A, Devel L, Georgiadis D, Hegele RG, Luo H, Granville DJ, Dive V, McManus BM, Overall CM. A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat Med. 2014;20:493–502. doi: 10.1038/nm.3508. [DOI] [PubMed] [Google Scholar]
- 73.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer. 2012;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- 75.Massagué J, Seoane J, Wotton D. Smad transcription factors. Gene Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 76.Min K-W, Liggett J, Silva G, Wu W, Wang R, Shen R-F, Eling TE, Baek SJ. NAG-1/GDF15 Accumulates in the nucleus and modulates transcriptional regulation of the Smad pathway. Oncogene. 2015 doi: 10.1038/onc.2015.95. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mishra S, Murphy LJ. Tissue Transglutaminase Has Intrinsic Kinase Activity: Identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase. J Biol Chem. 2004;279:23863–23868. doi: 10.1074/jbc.M311919200. [DOI] [PubMed] [Google Scholar]
- 78.Mishra S, Saleh A, Espino PS, Davie JR, Murphy LJ. Phosphorylation of Histones by Tissue Transglutaminase. J Biol Chem. 2006;281:5532–5538. doi: 10.1074/jbc.M506864200. [DOI] [PubMed] [Google Scholar]
- 79.Mu Y, Sundar R, Thakur N, Ekman M, Gudey SK, Yakymovych M, Hermansson A, Dimitriou H, Bengoechea-Alonso MT, Ericsson J, Heldin CH, Landstrom M. TRAF6 ubiquitinates TGF[beta] type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun. 2011;2:330. doi: 10.1038/ncomms1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mullen Alan C, Orlando David A, Newman Jamie J, Lovén J, Kumar Roshan M, Bilodeau S, Reddy J, Guenther Matthew G, DeKoter RP, Young Richard A. Master Transcription Factors Determine Cell-Type-Specific Responses to TGF-β Signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakamura T, Scorilas A, Stephan C, Yousef GM, Kristiansen G, Jung K, Diamandis EP. Quantitative analysis of macrophage inhibitory cytokine-1 (MIC-1) gene expression in human prostatic tissues. Br J Cancer. 2003;88:1101–1104. doi: 10.1038/sj.bjc.6600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oliverio S, Amendola A, Di Sano F, Farrace MG, Fesus L, Nemes Z, Piredda L, Spinedi A, Piacentini M. Tissue transglutaminase-dependent posttranslational modification of the retinoblastoma gene product in promonocytic cells undergoing apoptosis. Mol Cell Biol. 1997;17:6040–6048. doi: 10.1128/mcb.17.10.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park D, Choi S, Ha KS. Transglutaminase 2: a multi-functional protein in multiple subcellular compartments. Amino Acids. 2010;39:619–631. doi: 10.1007/s00726-010-0500-z. [DOI] [PubMed] [Google Scholar]
- 85.Park SH, Kim YS, Park BK, Hougaard S, Kim SJ. Sequence-specific enhancer binding protein is responsible for the differential expression of ERT/ESX/ELF-3/ESE-1/jen gene in human gastric cancer cell lines: Implication for the loss of TGF-beta type II receptor expression. Oncogene. 2001;20:1235–1245. doi: 10.1038/sj.onc.1204227. [DOI] [PubMed] [Google Scholar]
- 86.Parras-Molto M, Campos-Laborie F, Garcia-Dieguez J, Rodriguez-Grinolo M, Perez-Pulido A. Classification of protein motifs based on subcellular localization uncovers evolutionary relationships at both sequence and functional levels. BMC Bioinformatics. 2013;14:229. doi: 10.1186/1471-2105-14-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng X, Zhang Y, Zhang H, Graner S, Williams JF, Levitt ML, Lokshin A. Interaction of tissue transglutaminase with nuclear transport protein importin-α3. FEBS Lett. 1999;446:35–39. doi: 10.1016/s0014-5793(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 88.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012;31:2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 90.Prescott JD, Koto KS, Singh M, Gutierrez-Hartmann A. The ETS transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol Cell Biol. 2004;24:5548–5564. doi: 10.1128/MCB.24.12.5548-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quéré R, Karlsson G, Hertwig F, Rissler M, Lindqvist B, Fioretos T, Vandenberghe P, Slovak ML, Cammenga J, Karlsson S. Smad4 binds Hoxa9 in the cytoplasm and protects primitive hematopoietic cells against nuclear activation by Hoxa9 and leukemia transformation. Blood. 2011;117:5918–5930. doi: 10.1182/blood-2010-08-301879. [DOI] [PubMed] [Google Scholar]
- 93.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 94.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 95.Rudders S, Gaspar J, Madore R, Voland C, Grall F, Patel A, Pellacani A, Perrella MA, Libermann TA, Oettgen P. ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-kappa B to regulate the inducible nitric-oxide synthase gene. J Biol Chem. 2001;276:3302–3309. doi: 10.1074/jbc.M006507200. [DOI] [PubMed] [Google Scholar]
- 96.Sánchez-Tilló E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA. 2011;108:19204–19209. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Senapati S, Rachagani S, Chaudhary K, Johansson SL, Singh RK, Batra SK. Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene. 2010;29:1293–1302. doi: 10.1038/onc.2009.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shatnawi A, Norris JD, Chaveroux C, Jasper JS, Sherk AB, McDonnell DP, Giguere V. ELF3 is a repressor of androgen receptor action in prostate cancer cells. Oncogene. 2014;33:862–871. doi: 10.1038/onc.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shi Y, Massagué J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 101.Si-Tayeb K, Monvoisin A, Mazzocco C, Lepreux S, Decossas M, Cubel G, Taras D, Blanc JF, Robinson DR, Rosenbaum J. Matrix Metalloproteinase 3 Is Present in the Cell Nucleus and Is Involved in Apoptosis. Am J Pathol. 2006;169:1390–1401. doi: 10.2353/ajpath.2006.060005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in Inflammation and Cancer. Annual Review of Immunology. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 103.Sirover MA. Subcellular dynamics of multifunctional protein regulation: Mechanisms of GAPDH intracellular translocation. J Cell Biochem. 2012;113:2193–2200. doi: 10.1002/jcb.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith G, Carey FA, Beattie J, Wilkie MJV, Lightfoot TJ, Coxhead J, Garner RC, Steele RJC, Wolf CR. Mutations in APC, Kirsten-ras, and p53—alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song X, Wang X, Zhuo W, Shi H, Feng D, Sun Y, Liang Y, Fu Y, Zhou D, Luo Y. The Regulatory Mechanism of Extracellular Hsp90α on Matrix Metalloproteinase-2 Processing and Tumor Angiogenesis. J Biol Chem. 2010;285:40039–40049. doi: 10.1074/jbc.M110.181941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tatsukawa H, Fukaya Y, Frampton G, Martinez–Fuentes A, Suzuki K, Kuo TF, Nagatsuma K, Shimokado K, Okuno M, Wu J, Iismaa S, Matsuura T, Tsukamoto H, Zern MA, Graham RM, Kojima S. Role of Transglutaminase 2 in Liver Injury via Cross-linking and Silencing of Transcription Factor Sp1. Gastroenterology. 2009;136:1783–1795.e1710. doi: 10.1053/j.gastro.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Telci D, Wang Z, Li X, Verderio EAM, Humphries MJ, Baccarini M, Basaga H, Griffin M. Fibronectin-Tissue Transglutaminase Matrix Rescues RGD-impaired Cell Adhesion through Syndecan-4 and β1 Integrin Co-signaling. J Biol Chem. 2008;283:20937–20947. doi: 10.1074/jbc.M801763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsui KH, Hsu SY, Chung LC, Lin YH, Feng TH, Lee TY, Chang PL, Juang HH. Growth differentiation factor-15: a p53- and demethylation-upregulating gene represses cell proliferation, invasion, and tumorigenesis in bladder carcinoma cells. Sci Rep. 2015;5:12870. doi: 10.1038/srep12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Valenta T, Hausmann G, Basler K. The many faces and functions of [beta]-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Walker D, Poczobutt J, Gonzales M, Horita H, Gutierrez-Hartmann A. ESE-1 is Required to Maintain the Transformed Phenotype of MCF-7 and ZR-75–1 Human Breast Cancer Cells. Open Cancer J. 2010;3:77–88. [Google Scholar]
- 113.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, Tracey KJ. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 114.Wang JL, Chen ZF, Chen HM, Wang MY, Kong X, Wang YC, Sun TT, Hong J, Zou W, Xu J, Fang JY. Elf3 drives beta-catenin transactivation and associates with poor prognosis in colorectal cancer. Cell Death Dis. 2014;5:e1263. doi: 10.1038/cddis.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Baek SJ, Eling TE. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem Pharmacol. 2013;85:597–606. doi: 10.1016/j.bcp.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang X, Song X, Zhuo W, Fu Y, Shi H, Liang Y, Tong M, Chang G, Luo Y. The regulatory mechanism of Hsp90α secretion and its function in tumor malignancy. Proc Natl Acad Sci USA. 2009;106:21288–21293. doi: 10.1073/pnas.0908151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Y, Trepel JB, Neckers LM, Giaccone G. STA-9090, a small-molecule Hsp90 inhibitor for the potential treatment of cancer. Curr Opin Investig Drugs. 2010;11:1466–1476. [PubMed] [Google Scholar]
- 118.Wang YN, Lee HH, Lee HJ, Du Y, Yamaguchi H, Hung MC. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J Biol Chem. 2012;287:16869–16879. doi: 10.1074/jbc.M111.314799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang YN, Wang H, Yamaguchi H, Lee HJ, Lee HH, Hung MC. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem Biophy Res Comm. 2010;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Warner DR, Pisano MM, Roberts EA, Greene RM. Identification of three novel Smad binding proteins involved in cell polarity. FEBS Lett. 539:167–173. doi: 10.1016/s0014-5793(03)00155-8. [DOI] [PubMed] [Google Scholar]
- 121.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 122.Xu J, Acharya S, Sahin O, Zhang Q, Saito Y, Yao J, Wang H, Li P, Zhang L, Lowery Frank J, Kuo W-L, Xiao Y, Ensor J, Sahin Aysegul A, Zhang Xiang HF, Hung M-C, Zhang Jitao D, Yu D. 14–3–3ζ Turns TGF-β’s Function from Tumor Suppressor to Metastasis Promoter in Breast Cancer by Contextual Changes of Smad Partners from p53 to Gli2. Cancer Cell. 27:177–192. doi: 10.1016/j.ccell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yakubov B, Chen L, Belkin AM, Zhang S, Chelladurai B, Zhang ZY, Matei D. Small molecule inhibitors target the tissue transglutaminase and fibronectin interaction. PLoS One. 2014;9:e89285. doi: 10.1371/journal.pone.0089285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang H, Filipovic Z, Brown D, Breit SN, Vassilev LT. Macrophage inhibitory cytokine-1: a novel biomarker for p53 pathway activation. Mol Cancer Ther. 2003;2:1023–1029. [PubMed] [Google Scholar]
- 125.Yilmaz M, Christofori G. Mechanisms of Motility in Metastasizing Cells. Molecular Cancer Res. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 126.You H, Yamamoto K, Mak TW. Regulation of transactivation-independent proapoptotic activity of p53 by FOXO3a. Proc Natl Acad Sci USA. 2006;103:9051–9056. doi: 10.1073/pnas.0600889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhao Y, Aguilar A, Bernard D, Wang S. Small-Molecule Inhibitors of the MDM2–p53 Protein–Protein Interaction (MDM2 Inhibitors) in Clinical Trials for Cancer Treatment. J Med Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zimmers TA, Gutierrez JC, Koniaris LG. Loss of GDF-15 abolishes sulindac chemoprevention in the ApcMin/+ mouse model of intestinal cancer. J Cancer Res Clin Oncol. 2010;136:571–576. doi: 10.1007/s00432-009-0691-4. [DOI] [PubMed] [Google Scholar]
- 129.Zwijsen A, Verschueren K, Huylebroeck D. New intracellular components of bone morphogenetic protein/Smad signaling cascades. FEBS Lett. 2003;546:133–139. doi: 10.1016/s0014-5793(03)00566-0. [DOI] [PubMed] [Google Scholar]