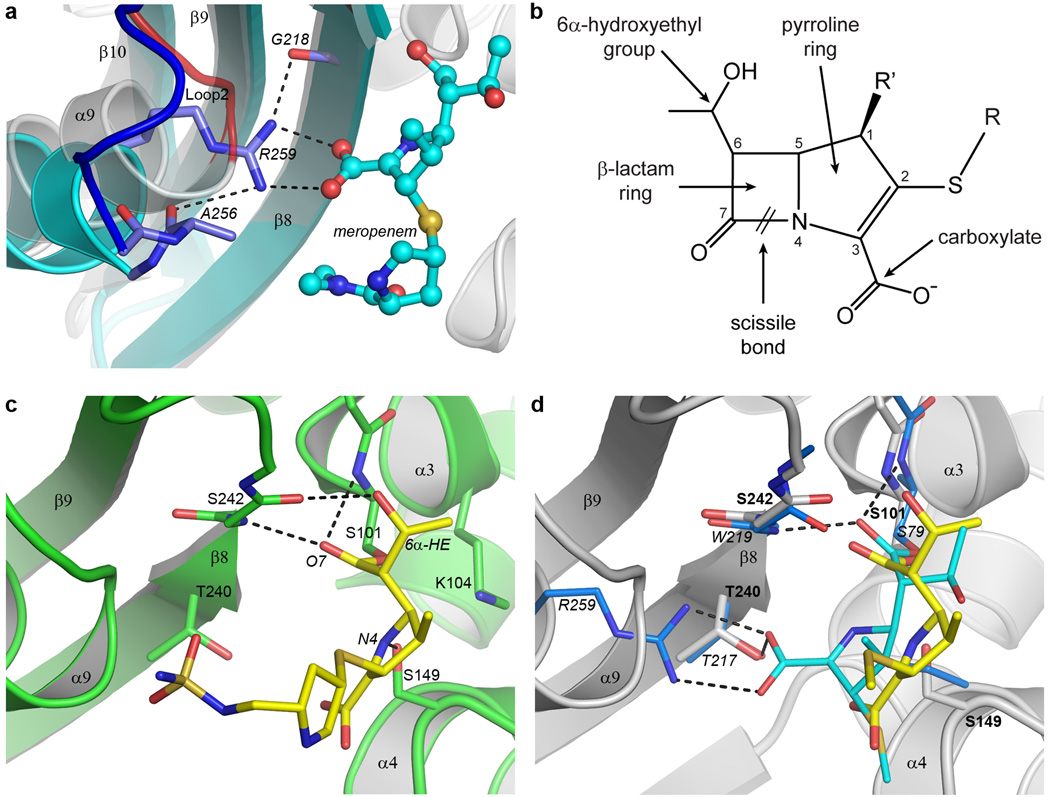
Figure 3. BPU-1 substrate binding.
(a) Close up view of the arginine pocket in OXA-23 (cyan ribbons, blue loop and lighter blue sticks), superimposed on the equivalent region near Loop 2 in BPU-1 (gray ribbons and red loop). The conserved Gram-negative class D arginine residue (represented by Arg259 from OXA-23) responsible for anchoring the carboxylate group of meropenem (cyan ball-and-stick) in OXA-23 is indicated. (b) The general structure of the carbapenem antibiotics. Differences in the structure of the tail group (R) distinguish the various clinically-available antibiotics including imipenem, meropenem, doripenem, and ertapenem. The R’ position is occupied either by hydrogen in imipenem) or by a methyl group in meropenem, doripenem, and ertapenem. (c) The substrate binding site in the doripenem-BPU-1 complex. The BPU-1 structure is represented by green ribbons and sticks and the covalently-linked doripenem by yellow sticks. The hydrogen bonding interactions between the doripenem and the protein are shown as dashed black lines. (d) The difference in coordination of doripenem (yellow sticks) in BPU-1 (grey ribbons, grey sticks) and meropenem (cyan sticks) in OXA-23. Only the residues which interact with the meropenem are shown for OXA-23 (light blue sticks) for clarity, and only the hydrogen bonding in the meropenem-OXA-23 complex is shown. The OXA-23 residues are labeled in italics and the BPU-1 residues in plain bold text. The tail of the doripenem in BPU-1 is truncated at the sulfur atom for clarity.