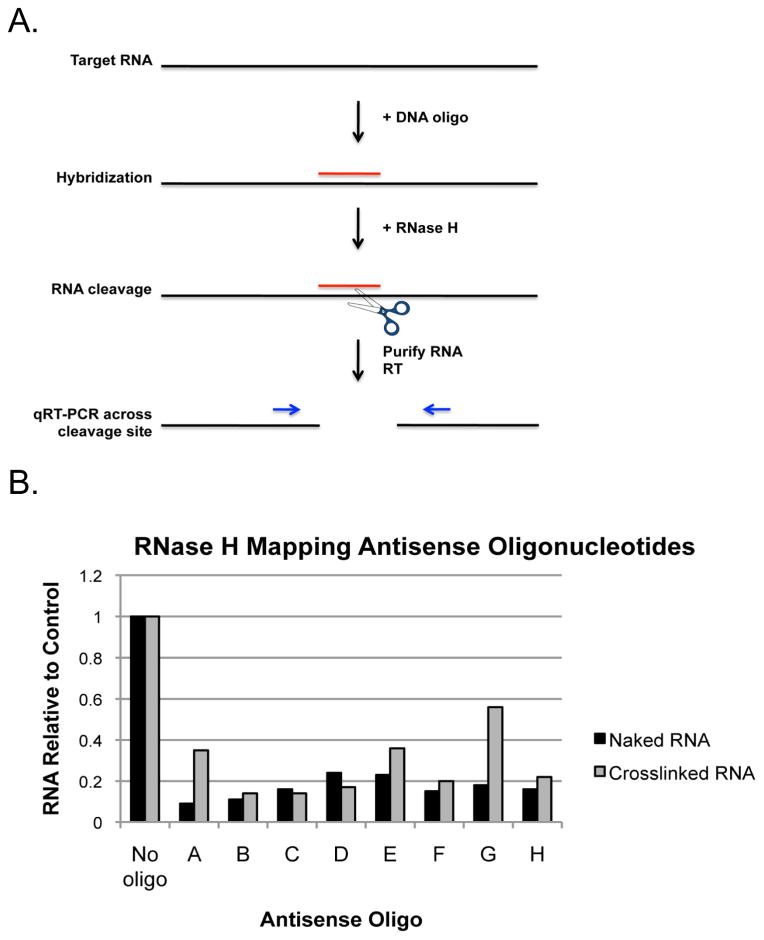
Figure 2. RNase H mapping antisense oligonucleotide positions.
A) Schematic of the RNase H mapping assay. Target RNA is incubated with complementary antisense DNA oligonucleotides and RNase H. If hybridization occurs, RNase H will cleave the RNA involved in the RNA:DNA hybrid. Cleavage is measured using RT-qPCR. B) Infected cells were cross-linked, lysed, and incubated with candidate antisense DNA oligonucleotides and RNase H. Purified RNA from mock cross-linked cells was used as a positive control for RNase H cleavage. RNA was purified from RNase H reactions and cleavage assessed by RT-qPCR using qPCR primers that amplify across the oligonucleotide hybridization site. Data is represented as the amount of RNA remaining relative to the no oligo control.