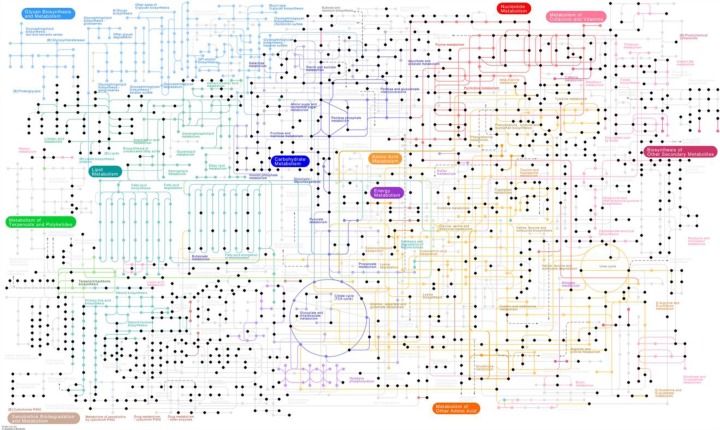
Figure 2.
Example coverage of human metabolism by a high-resolution metabolomics experiment. From a set of human plasma samples, we generated the data set via a Thermo Fisher Q Exactive mass spectrometer, with reverse-phase C18 liquid chromatography and negative ionization. The experiment produced 35,708 metabolite features, which were then matched to the KEGG pathway database within 2 parts per million mass accuracy. Data from this experiment match to 879 metabolites in the KEGG human metabolic map. Each black dot represents a tentatively matched metabolite. Light gray dots are from other species and are not considered as part of human metabolism by KEGG. Complemented by other ionization methods and chromatographic tools, the practical coverage will be further increased.