Abstract
Competence is a physiological state that enables Bacillus subtilis 168 to take up and internalize extracellular DNA. In practice, only a small subpopulation of B. subtilis 168 cells becomes competent when they enter stationary phase. In this study, we developed a new transformation method to improve the transformation efficiency of B. subtilis 168, specially in rich media. At first, different competence genes, namely comK, comS, and dprA, were alone or together integrated into the chromosome of B. subtilis 168 under control of mannitol-inducible PmtlA promoter. Overexpression of both comK and comS increased the transformation efficiency of B. subtilis REG19 with plasmid DNA by 6.7-fold compared to the wild type strain 168. This transformation efficiency reached its maximal level after 1.5 h of induction by mannitol. Besides, transformability of the REG19 cells was saturated in the presence of 100 ng dimeric plasmid or 3000 ng chromosomal DNA. Studying the influence of global regulators on the development of competence pointed out that important competence development factors, such as Spo0A, ComQXPA, and DegU, could be removed in REG19. On the other hand, efficient REG19 transformation remained highly dependent on the original copies of comK and comS regardless of the presence of PmtlA-comKS. Finally, novel plasmid-free strategies were used for transformation of REG19 based on Gibson assembly.
Keywords: DNA uptake, transformation rate, competence, mannitol induction, comK
Introduction
Since the first report on transformation of Bacillus subtilis by Spizizen (1958), the competency of B. subtilis has intensively been studied revealing highly interconnected regulatory networks (Hamoen et al., 2003). These interconnected regulatory networks are responsible for fine-tuning of the single-cell and social behavior of B. subtilis in a culture medium, specially during their entrance to stationary phase. When B. subtilis cells enter stationary phase due to nutrient deprivation and high cell density, they start to differentiate into various subpopulations. Some of them become motile (Nishihara and Freese, 1975), while the others form biofilm (Vlamakis et al., 2008), secrete degradative enzymes and antibiotics (González-Pastor et al., 2003), or finally sporulate (Rudner and Losick, 2001; Piggot and Hilbert, 2004). Another small subpopulation differentiates into competent cells able to take up extracellular DNA (Dubnau, 1991a; Dubnau and Provvedi, 2000).
Generally, the main actor in B. subtilis differentiation processes is Spo0A, the transcriptional master regulator of sporulation. Spo0A is phosphorylated via a complex phosphorelay system consisting of five histidine kinases (KinA, B, C, D, and E), a phosphate acceptor protein (Spo0F) and a phosphotransferase (Spo0B) (Burbulys et al., 1991). Depending on the level of phosphorylated Spo0A, the cells individually enter different states. Specifically, B. subtilis becomes competent when the competence transcription factor, ComK, reaches a certain threshold level (Maamar and Dubnau, 2005; Smits et al., 2005). ComK is the competence master regulator which activates about 100 genes for DNA-recombination, -repair, -binding, -uptake (Berka et al., 2002; Hamoen et al., 2002), cell division (Hamoen, 2011), as well as its own promoter in a positive feedback loop (van Sinderen and Venema, 1994). Therefore, the ComK pool in the cell is tightly regulated at the transcriptional level by the antagonizing global regulators AbrB (the transition state regulator) and Spo0A, CodY, and its specific regulator Rok (Hoa et al., 2002), as well as the DegS–DegU two component system (Hamoen et al., 2000; Figure 1).
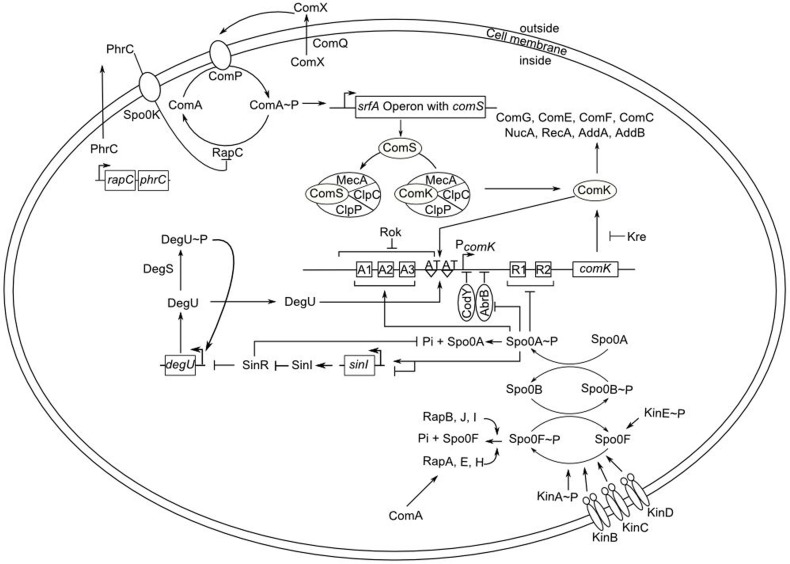
Figure 1.
Overview of competence regulation in B. subtilis. Lines ending in perpendiculars and arrows denote negative and positive effects, respectively. The quorum sensing mechanism containing the modified peptide ComX results in phosphorylation of the ComA response regulator by ComP. ComA~P activates the transcription of the srf operon with the embedded comS gene. ComS binds to the protease complex of MecA, ClpC, and ClpP and prevents the degradation of the transcription factor ComK. The stability of the comK mRNA is also influenced by Kre. The transcription of comK is regulated by various factors. ComK activates its own expression by binding to the AT-boxes in its promoter region. This binding is facilitated by DegU. At low concentration, Spo0A~P binds to three special sequences (A1–A3; high affinity binding sites) in the promotor of comK. At high concentration, Spo0A~P is capable of binding to further sequences (R1 and R2; low affinity binding sites) and repressing the expression of comK. Spo0A~P also release the comK expression by repressing AbrB. Other repressing factors of the comK expression are CodY and Rok. Rap proteins, such as RapA, prevent ComA~P interaction with its target DNA and dephosphorylate Spo0F~P. The expression of sinI is activated at the low level of Spo0A~P and repressed by a high level of Spo0A~P concentration. SinI antagonizes SinR, which represses the expression of degU. Finally, high concentration of ComK activates the expression of genes for DNA-uptake and -integration as well as many other genes.
During exponential phase, the expression of comK is repressed by Rok, AbrB, and by CodY, a pleiotropic repressor responding to the GTP level in the cell (Serror and Sonenshein, 1996). Recently, it has been found that the stability of comK mRNA is also influenced by Kre (YkyB) (Gamba et al., 2015; Figure 1). Moreover, the ComK pool in the cell is regulated at the posttranslational level. ComK is targeted by the adaptor protein MecA and degraded in a complex with ClpC and ClpP (Kong and Dubnau, 1994; Turgay et al., 1997). At the end of the exponential phase, changing the levels of DegU and phosphorylated Spo0A drastically alter the situation enabling the expression of comK. Briefly, DegU is phosphorylated by DegS in response to unknown intracellular signals. While phosphorylated DegU regulates the synthesis of degradative enzymes, biofilm formation, and capsule synthesis, DegU binds to the comK promoter region, thereby activates the transcription of comK (Dahl et al., 1992; Figure 1). Moreover, phosphorylated Spo0A represses the expression of abrB, releasing comK expression from repression by AbrB (Hahn et al., 1995). Therefore, Δspo0A strains are poorly transformable (Sadaie and Kada, 1983; Albano et al., 1987). In a concerted manner with the relief of comK from repression, a small protein, ComS, is able to protect ComK from the ClpC-ClpP-MecA complex during the stationary phase (Turgay et al., 1998; Prepiak et al., 2011; Figure 1). The comS gene is located within the srf open reading frame encoding surfactin and its expression is regulated by quorum sensing signaling pathway provoked by the ComQXPA cluster. ComX, an extracellular pheromone modified by ComQ, is sensed by a membrane-bound sensor histidine kinase (ComP), eventually resulting in the activation of its cognate response regulator ComA (Magnuson et al., 1994; Comella and Grossman, 2005). Phosphorylated ComA induces expression of comS and activates natural competence (Hahn and Dubnau, 1991; Figure 1).
When ComK molecules reach a sufficient amount and the cells become competent, they are able to take up DNA. On the surface of a competent cell, the double-stranded DNA binds to a receptor via a pseudopilus similar to type IV pili, cleaved by a membrane-localized nuclease and internalized as a linear single stranded DNA (ssDNA). Inside the cell, ssDNA is protected by single strand DNA binding proteins, such as DprA, SsbA, SsbB, and RecN (Inamine and Dubnau, 1995). During transformation, DprA protects the DNA inside the cell and is more important during the transformation with plasmid than with chromosomal DNA (Tadesse and Graumann, 2007). Moreover, DprA facilitates the displacement of SsbA and SsbB and mediates the attachment of RecA to ssDNA, which has already been covered by Ssb proteins (Yadav et al., 2013). Finally, the ssDNA is integrated into the genome via homologous recombination by RecA (Kidane et al., 2012).
Despite all the theoretical progress, B. subtilis is routinely transformed using a minimal medium according to the 50 years old method described by Anagnostopoulos and Spizizen (1961), albeit with slight modifications. This method is a time-consuming and inefficient method for transformation, specially for natural B. subtilis isolates (Duitman et al., 2007). To improve transformation efficiency, several attempts were carried out with regulated expression of comK as well as comS (Hahn et al., 1996; Liu et al., 1996; Duitman et al., 2007; Nijland et al., 2010; Zhang and Zhang, 2011). In this work, we expressed comK, comS, and dprA alone or in combined artificial operons under control of the B. subtilis mannitol promoter (PmtlA). In this way, we studied the influence of other important factors like ComQXPA, DegU, and Spo0A on transformation efficiency of plasmids and chromosomal DNA in these strains. Thereby, we constructed a strain with high transformation efficiency in rich medium. To our knowledge, this is the first attempt for overexpression of two competence factors in combination.
Materials and methods
Strains, media, and growth condition
All bacterial strains used in this study are listed in Table 1. Escherichia coli JM109 was used for plasmid construction and propagation. E. coli strain NM538 was used for multimerization of the monomer plasmids. All E. coli transformants were selected on LB agar (Bertani, 1951) supplemented with ampicillin (100 μg ml−1) or spectinomycin (100 μg ml−1) depending on the plasmid marker. When plasmids were used for transformation of B. subtilis, transformants were selected on LB agar containing kanamycin (10 μg ml−1), spectinomycin (100 μg ml−1), erythromycin (5 μg ml−1), or chloramphenicol (5 μg ml−1). The tryptophan or histidine auxotroph mutants were cultivated on Spizizen's minimal medium (Anagnostopoulos and Spizizen, 1961) supplemented with 50 μg ml−1 tryptophan or histidine, respectively. B. subtilis strains transformed with chromosomal DNA of strain KM0 (trp+) were selected on Spizizen's minimal medium containing glucose (0.5% w/v).
Table 1.
Strains used in this study.
| Strain | Genotype | Source, Reference, or Construction |
|---|---|---|
| E. coli | ||
| JM109 | recA1, endA1, gyrA96, thi-1, hsdR17(, ), mcrA, supE44, gyrA96, relA1, λ−, Δ(lac-proAB), F′ (traD36, proAB+, lacIq, lacZΔM15) | Yanisch-Perron et al., 1985 |
| NM538 | supF, hsdR, trpR, lacY | Frischauf et al., 1983 |
| B. subtilis | ||
| 168 | trpC2 | DSM23778* |
| IIG-Bs168-1 | trpC2, ΔmanPA::ermC | pJOE6577.1 → 168 |
| KM0 | trpC | pKAM041 → 168 |
| REG1 | trpC2, ΔmanPA::ermC, hisI′-spcR-yvcA | pHM30 → IIG-Bs168-1 |
| REG3 | trpC2, ΔmanPA::ermC, PmtlA-comS | pREG3 → REG1 |
| REG4 | trpC2, ΔmanPA::ermC, PmtlA-dprA | pREG4 → REG1 |
| REG5 | trpC2, ΔmanPA::ermC, PmtlA-dprA-comS | pREG5 → REG1 |
| REG6 | trpC2, ΔmanPA::ermC, PmtlA-comK | pREG6 → REG1 |
| REG7 | trpC2, ΔmanPA::ermC, PmtlA-comK-dprA | Gibson 1 → REG1 |
| REG19 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS | pJOE7361.1 → REG1 |
| REG32 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS, ΔcomS | pREG12 → REG19 |
| REG33 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS, ΔcomK | pREG13 → REG19 |
| REG35 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS, ΔcomK, ΔcomS | pREG13 → REG32 |
| REG36 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS, Δspo0A | pJOE7122 → REG19 |
| REG37 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS, ΔcomQXPA | pREG10 → REG19 |
| REG120 | trpC2, ΔmanPA::ermC, PmtlA-comK-comS, ΔdegU | pREG58 → REG19 |
Deutsche Sammlung von Mikroorganismen und Zellkulturen.
DNA manipulation
Oligonucleotides used in this study were synthesized by Eurofins MWG Operon (Ebersberg, Germany; Table S1). All plasmids used in this study are listed in Table 2. Standard molecular techniques were carried out as described before (Sambrook et al., 1989). Chemicals, enzymes, and kits used for this study are described in Supplementary Materials.
Table 2.
Plasmids used in this study.
| Plasmid | Genotype | Source or Reference |
|---|---|---|
| pHM30 | oripUC18, bla, ′hisF-hisI′-spcR-′yvcA-yvcB′ | Motejadded and Altenbuchner, 2007 |
| pHM31 | oripUC18, bla, ′hisF-hisI-′yvcA-yvcB′ | Motejadded and Altenbuchner, 2007 |
| pIC20R | oripUC18, bla, lacZα | Marsh et al., 1984 |
| pJOE2962 | oripACYC184, cat, rhaPBAD-mrpA | Warth et al., 2011 |
| pJOE4370.7 | ori+pUB110, oripUC18, reppUB110, kanR, bla, ble | Altenbuchner, unpublished |
| pJOE4786.1 | oripUC18, bla, ter-′lacI-lacZα-ter | Altenbuchner et al., 1992 |
| pJOE6577.1 | oripUC18, bla, spcR, ′manR-manP-ermC-yjdF | Wenzel and Altenbuchner, 2015 |
| pJOE6743.1 | oripUC18, spcR, bla, PmanP-manP | Wenzel and Altenbuchner, 2015 |
| pJOE6905.1 | oripUC18, bla, ter-PmtlA-comK-ter | This study |
| pJOE7122.1 | oripUC18, spcR, bla, PmanP-manP, yqiG-recN′ | This study |
| pJOE7331.2 | oripUC18, bla, PmtlA-comK-comS | This study |
| pJOE7361.1 | oripUC18, bla, yvcB′-PmtlA-comK-comS-hisI-hisF′ | This study |
| pKAM041 | oripUC18, bla, ter-′trpD-trpC-trpF′-ter | This study |
| pKAM180 | oripUC18, bla, ′hisF-hisI-ter-luc-PmtlA-ter-′yvcA-yvcB′ | This study |
| pREG3 | oripUC18, bla, yvcB′-PmtlA-comS-hisI-hisF′ | This study |
| pREG4 | oripUC18, bla, yvcB′-PmtlA-dprA-hisI-hisF′ | This study |
| pREG5 | oripUC18, bla, yvcB′-PmtlA-dprA-comS-hisI-hisF′ | This study |
| pREG6 | oripUC18, bla, yvcB′-PmtlA-comK-hisI-hisF′ | This study |
| pREG10 | oripUC18, spcR, bla, PmanP-manP,′mrpG-degQ | This study |
| pREG12 | oripUC18, spcR, bla, PmanP-manP, ′hxlR-ycxA′ | This study |
| pREG13 | oripUC18, spcR, bla, PmanP-manP, ′yhzC-yhxD′ | This study |
| pREG58 | oripUC18, spcR, bla, PmanP-manP, ′degS-yviA′ | This study |
| pWAL275 | ori+pUB110, oripUC18, reppUB110, kanR, bla, mrpS | Warth and Altenbuchner, unpublished |
Construction of the integration plasmid carrying functional trpC
To facilitate the growth of B. subtilis 168 in minimal media, the trpC2 gene was replaced with functional trpC using the pKAM041 integration plasmid. The deleted codon was added to the trpC2 sequence using primary PCRs with s8475-s8634 and s8633-s8476 oligonucleotide pairs. The primary PCR products were then fused in a PCR using oligonucleotides s8475 and s8476. The final PCR product was then inserted into pJOE4786.1 cut with SmaI to create pKAM041.
Construction of integration plasmids for inducible gene expression
To integrate the PmtlA-comK cassette into the B. subtilis chromosome, the PmtlA DNA fragment was amplified in a PCR using oligonucleotides s6851 and s6853. In the second PCR, the comK gene together with its ribosomal binding site was amplified with oligonucleotides s6854 and s6852. The two primary PCR products overlapping each other were finally fused in a PCR with primers s6851-s6852. The PmtlA-comK DNA fragment was then inserted into the pJOE4786.1 plasmid via SmaI sites creating pJOE6905.1. Next, the PmtlA-comK cassette was cut out from pJOE6905.1 by BamHI and inserted into pHM31 via the same restriction site to create pREG6. For generation of the PmtlA-comK-comS (or PmtlA-comKS) cassette, the PmtlA-comK DNA fragment was amplified from pJOE6905.1 in a PCR using oligonucleotides s6851 and s7707. Primers s7708 and s7709 were used to amplify the comS gene from B. subtilis 168 in the second PCR. Both PmtlA-comK and comS DNA fragments were fused in a PCR with oligonucleotides s6851 and s7709. The PCR product (PmtlA-comKS) was primarily inserted into pJOE4786.1 via SmaI sites (pJOE7331.2). The PmtlA-comKS cassette was then cut out from pJOE7331.2 by BamHI and inserted into pHM31 through BamHI sites creating pJOE7361.1.
Further integration plasmids for inducible gene expression were constructed based on pKAM180 (for construction see Supplementary Materials), another pHM31-derivative containing PmtlA-luc (luciferase) cassette. Both comS and dprA were amplified with their ribosomal binding site using oligonucleotide pair s8003-s8004 (comS) and s8005-s8006 (dprA), respectively, for creating the PmtlA-comS and PmtlA-dprA cassettes. Each PCR fragment was then inserted via AflII and NheI into pKAM180 to create pREG3 (PmtlA-comS) and pREG4 (PmtlA-dprA). For generation of the PmtlA-dpA-comS, the amplified comS DNA fragment was cut by BrsGI and AflII, whereas the PCR fragment of dprA by BsrGI and NheI. Afterwards, both fragments were inserted into pKAM180 digested with AflII and NheI to create pREG5. For generation of the PmtlA-comK-dprA, the amplified dprA DNA fragment with the ribosomal binding site was amplified using oligonucleotide pair s10576-s10577 with overlapping sequences to pREG6. pREG6 was cut by NheI and the PCR fragment of dprA integrated by Gibson Assembly as described below (Gibson 1).
Construction of gene deletion plasmids
Deletion plasmids were mainly constructed based on pJOE6743.1 carrying spectinomycin resistance gene (selection marker) and PmanP-manP (anti-selection marker). Each deletion cassette was created by ligation of the two flanking fragments (cut EcoRI and SphI) of the target deletion region with pJOE6743.1 (cut SphI) in a three fragment ligation. For the comQXPA deletion, the oligonucleotide pair s8125–s8126 and s8127–s8128 were used and the PCR fragments were inserted via SphI and EcoRI (pREG10). For the comS deletion, oligonucleotides s8172–s8249 and s8250–s8175 were used and inserted via BamHI and EcoRI (pREG12). Oligonucleotides s8251–s8132 and s8252–s8253 were used for the comK deletion and inserted via SphI and EcoRI (pREG13). Deletion of degU was carried out using oligonucleotides s9314–s9315 and s9316–s9317 followed by the insertion of the PCR products via SphI and EcoRI into pJOE6743.1 (pREG58). For the spo0A-spoIVB deletion, oligonucleotides s7328–s7329 and s7330–s7331 were used. The amplified fragments were fused in a PCR and inserted blunt ended between the SmaI sites of pJOE6743.1 (pJOE7122.1).
Construction and multimerization of the pWAL275 plasmid
The pJOE4370.7 plasmid is a shuttle vector consisting of a 3.7 kb ScaI-PvuII fragment of pUB110 inserted into the SmaI site of plasmid pIC20R. To create pWAL275, the mrpS site for site-specific recombination of plasmid SCP2 was inserted between AatII and HindIII sites of pJOE4370.7 using two complementary oligonucleotides mrpS-A and mrpS-B. Transformation of E. coli JM109 containing the mrpA-expressing pJOE2962 with the pWAL275 monomers led to multimerization of pWAL275. Plasmid DNA was then isolated from JM109 pJOE2962 pWAL275 (chloramphenicol and ampicillin resistance) again and used to transform JM109 without pJOE2962 (ampicillin resistance). Finally, the transformants were screened for the pWAL275 dimers using agarose gel electrophoresis (Warth et al., 2011). In all experiments, the dimers of pWAL275 were used for transformation of B. subtilis strains.
Bacterial transformation
Transformation of E. coli with plasmid DNA was done as described previously (Chung et al., 1989). The “Paris method” was used to transform B. subtilis strains in Spizizen's minimal medium as described before (Harwood and Cutting, 1990). To transform the constructed strains carrying competence genes (comK, comS, dprA) expressed by the B. subtilis mannitol promoter (PmtlA), a new transformation protocol was developed. A 5 ml overnight culture, inoculated from a single colony, with an OD600 of about 1.7 was used to inoculate shake flasks containing 10 ml LB with an OD600 of 0.1 and incubated at 37°C with 200 rpm. After 90 min incubation, 0.5% (w/v) mannitol was added and the bacterial culture was further incubated for 90 min. The cells were then washed in LB medium and diluted to an OD600 of 0.5. Unless otherwise stated, 100 ng of the pWAL275 dimers or 3 μg of chromosomal DNA of the KM0 strain were added to each 1 ml aliquots of the cell suspension. After adding the desired DNA, the cell suspension was incubated on a roller drum for 1 h at 37°C. Depending on the DNA, transformants were selected as described above.
Construction of B. subtilis strains by markerless gene deletion/integration systems
In this study, two strains, KM0 (trp+) and IIG-Bs168-1 (trpC2 ΔmanPA), were constructed by direct transformation of B. subtilis 168 (trpC2). Primarily, strain 168 was transformed with pKAM041 in Spizizen's minimal medium containing tryptophan (50 μg ml−1) in order to replace the trpC2 mutation with the functional trpC gene. The 168 transformants with pKAM041 were selected on Spizizen's minimal medium without tryptophan. In this way, the tryptophan prototroph strain KM0 was constructed. On the other hand, transformation of strain 168 with pJOE6577.1 created strain IIG-Bs168-1, the parental host for the markerless gene deletion system. In IIG-Bs168-1, the manPA operon encoding mannose-specific PTS transporter and mannose 6-phosphate dehydrogenase were replaced by an erythromycin resistance gene. The manPA deletion was the essential modification for the markerless gene deletion method in B. subtilis using pJOE6743.1 derivatives (Wenzel and Altenbuchner, 2015). Markerless integration of the desired promoter-gene cassettes, such as PmtlA-comK, was carried out using the pHM30/pHM31 system based on histidine auxotrophy (Motejadded and Altenbuchner, 2007). Briefly, first the linearized plasmid pHM30 was integrated into the IIG-Bs168-1 (his+) genome via homologous recombination replacing the hisI gene with a spectinomycin resistance gene (strain REG1). In the second step, the his-auxotophic strain REG1 was transformed by a linearized suicide vector containing the hisI gene together with the PmtlA-gene cassette and selected for histidine-prototrophic and spectinomycin sensitive colonies.
Gene deletion using cassettes constructed by Gibson assembly
Three PCR fragments were fused using the Gibson Assembly® Cloning Kit (New England BioLabs®, Frankfurt, Germany) as recommended by the manufacturer to generate a linear DNA cassette for disruption of the amyE gene in B. subtilis (encodes α-amylase). The upstream homolog fragment of amyE was amplified in a PCR using oligonucleotides s10446 and s10447, while the downstream homolog fragment was amplified with oligonucleotides s10450 and s10451. Between the amplified upstream and downstream flanking fragments, the spectinomycin resistance gene (spcR) was amplified with oligonucleotides s10448 and s10449. These three DNA fragments were assembled to create the deletion cassette. In a second strategy, the flanking fragments of the deletion cassette were amplified using oligonucleotides s10453–s10447 and s10450–s10452. These newly amplified flanking fragments contained 20 base pairs overlap at their outer ends enabling them to form multimers or circulated DNA fragments after Gibson assembly. After fragments assembly, 10 μl of each mixture (single linear vs. multimer linear and circular cassettes) were used for REG19 transformation.
Results
Induction of competence by insertion of comK under control of pmtlA
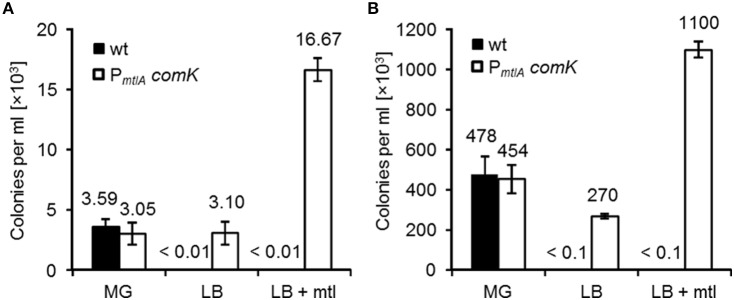
ComK is the master regulator of competence in B. subtilis. In practice, competence in B. subtilis is induced in minimal medium by quorum sensing signals and nutritional stress. After extensive growth in an amino acid rich medium, the cells are shifted to an amino acid deprived medium for a short time to induce competence in a two-step procedure (Paris method). To render competence independent of the growth medium and to enhance the transformation efficiency, the comK gene was inserted under control of the B. subtilis mannitol promoter (PmtlA) and integrated into the chromosome of B. subtilis 168 (strain REG6). Likewise, a new protocol with Luria–Bertani (LB) medium was developed to transform REG6. Briefly, an overnight culture was diluted in LB and incubated for 90 min at 37°C on a rotary shaker. Afterwards, mannitol was added at a final concentration of 0.5% and the culture was further incubated for 90 min. Cells were harvested, washed and resuspended in 1 ml aliquots. DNA was then added and cells were plated on selective agar plates after 1 h incubation at 37°C. To compare the transformation efficiency between B. subtilis 168 (wt) and REG6, the Paris method and the new protocol were used. The wt and REG6 strains were transformed using either pWAL275 dimers (a pUB110-pUC18 derivative) or chromosomal DNA of a trpC+ derivative of B. subtilis 168 (strain KM0). In the case of pWAL275, the transformed cells were selected on LB with kanamycin, whereas in the case of chromosomal DNA, the transformed cells were selected on Spizizen's minimal agar plates without tryptophan. The results are shown in Figures 2A,B. There was no significant difference between transformation efficiency of the wt and REG6 using plasmid DNA (Figure 2A) or chromosomal DNA (Figure 2B) when the Paris method with Spizizens' minimal glucose (or MG medium) was used. In LB medium without mannitol, very few kanamycin resistant and trpC+ colonies were obtained with the wild type strain. In strain REG6, the number of transformants was significantly increased in LB. When REG6 cells in LB were induced with mannitol, the number of transformants was increased by about 2.3-fold compared to the Paris method using chromosomal DNA and 4.6-fold using plasmid DNA. Transformation under DNA saturation condition with chromosomal DNA was about 100-fold more efficient than with plasmid DNA.
Figure 2.
Transformation efficiency of B. subtilis 168 (wt) and REG6 (PmtlA-comK). Cells were transformed according to Paris method using Spizizen's minimal medium containing glucose (MG) and the new protocol using LB medium with(out) mannitol. 100 ng of the pWAL275 dimers (A) and 3 μg of KM0 chromosomal DNA (B) were used for transformation. Error bars represent standard deviation from the mean value of triple experiments.
Enhancing transformation efficiency by induction of comK, dprA, and comS
Induction of comK by mannitol resulted in a higher transformation efficiency; however, competence development depends on more factors than ComK. For example, ComS controls degradation of ComK, and DprA conveys incoming ssDNA to RecA. To investigate the effect of ComS and DprA on transformation efficiency, comS and dprA alone, together or combined with comK were inserted downstream of PmtlA and integrated into the B. subtilis 168 chromosome. The new strains were called REG3, REG4, REG5, REG7, and REG19. To compare these strains with REG6, all strains were grown in LB, induced with mannitol and transformed with pWAL275 dimer DNA (Figure 3). Induction of comS (REG3) and dprA (REG4) alone increased the number of transformants in comparison with the wt, albeit less efficient than REG6. The concomitant expression of dprA and comS (REG5) as well as dprA and comK (REG7) showed more transformants than with REG4 even though the number of transformed cells remained less than with REG6. Interestingly, REG19 with comK and comS combined in an artificial operon was superior to all other strains. The plasmid transformation frequency of REG19 was about 6.7-fold higher in comparison with wt transformed in MG medium. Hereafter, transformation efficiency of REG19 was studied in more detail.
Figure 3.
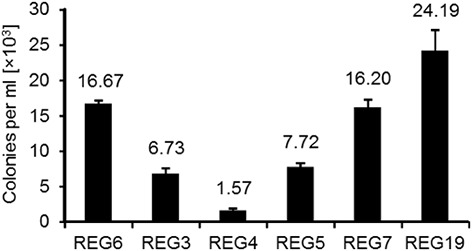
Overexpression of different competence genes and their effect on transformation efficiency. Strains B. subtilis REG6 (PmtlA-comK), REG3 (PmtlA-comS), REG4 (PmtlA-dprA), REG5 (PmtlA-dprA-comS), REG7 (PmtlA-comK-dprA), and REG19 (PmtlA-comK-comS) were transformed with 100 ng pWAL275 dimers in LB with mannitol as described in Section Materials and Methods. Error bars represent standard deviation from the mean value between triple experiments.
Optimization of the transformation protocol
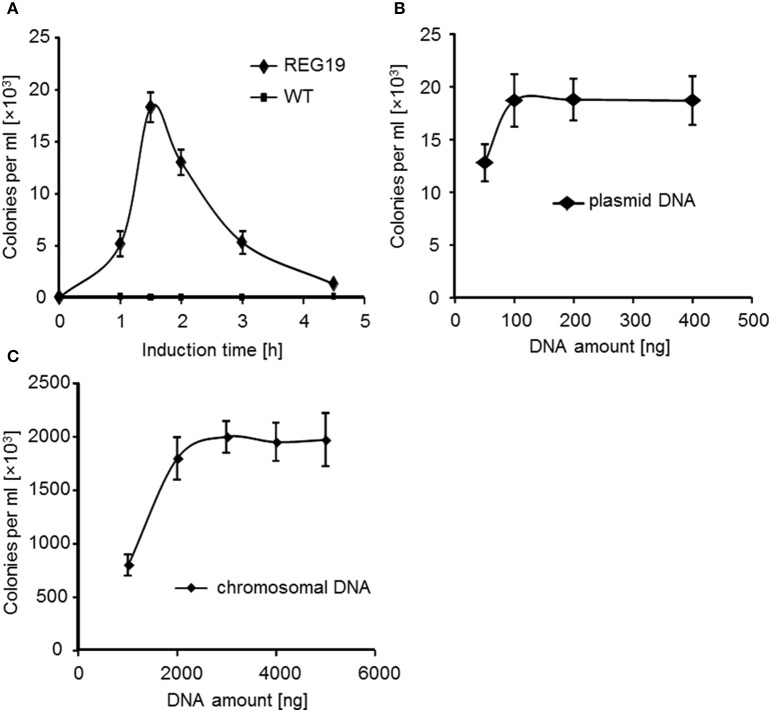
As noted above, the simultaneous induction of comK and comS resulted in the strongest transformation efficiency of B. subtilis. To see, whether the growth condition could be further optimized for competence development, the time of induction with mannitol was varied. As before, an overnight culture of the REG19 strain in LB was diluted in fresh LB, incubated in shake flasks for 1.5 h at 37°C and 0.5% (w/v) mannitol was added. Cultures were grown for 1, 1.5, 3, or 4.5 h to allow the induction of competence. The induced cultures were washed and diluted to an OD600 of 0.5 in 1 ml LB. pWAL275 DNA was added and the cultures were further incubated at 37°C for 60 min. Finally, dilutions were plated on LB agar containing kanamycin and the number of transformants counted at the next day. As shown in Figure 4A the highest number of transformants was achieved after 1.5 h of induction. Induction shorter or longer than 1.5 h decreased the transformation efficiency.
Figure 4.
Transformation efficiency of REG19 in LB medium with mannitol under different conditions. (A) Transformation of strains 168 (wt), as a control, and REG19 was carried out with 100 ng pWAL275 dimers after different time of induction with mannitol. (B) Transformation efficiency of REG19 was also studied after 1.5 h of induction with 50–400 ng of dimer plasmid DNA or (C) 1000–5000 ng of chromosomal DNA. Error bars represent standard deviation from the mean value between triplicate experiments.
Variable DNA samples and amount, e.g., 50–400 ng DNA of the pWAL275, were added to REG19 in order to determine the transformation efficiency (Figure 4B). The correlation between transformation efficiency and DNA amount showed that there is DNA saturation point at 100 ng of dimer plasmid DNA. At DNA saturation condition, 19 × 103 kanamycin resistant colonies per 1 ml competent cells were obtained with 100 ng pWAL275 dimer. 100 ng of the pWAL275 dimer equals 8 × 109 DNA molecules. Since the OD600 of 0.5 corresponds to 2.5 × 108 cells, the proportion of plasmid molecules to cells needed for achieving the maximal transformation rate is 32-fold for the dimer.
Further transformation experiments were done with REG19 and chromosomal DNA of B. subtilis KM0, by selection of the trpC+ transformants. In this case, saturation of transformation was obtained at about 3 μg chromosomal DNA (Figure 4C). Compared to plasmid DNA, the number of transformants was about 100- to 200-fold higher (2 × 106 cells).
The importance of original comS and comK in REG19
So far, we have shown that the induction of combined comK and comS (REG19) results in high transformation efficiency of B. subtilis. To understand whether the synthesis of ComK and ComS driven by PmtlA can bypass the requirements for regular competence-inducing factors, original comK and comS were deleted in REG19. For these experiments, strains REG33 (REG19 ΔcomK), REG32 (REG19 ΔcomS) and REG35 (REG19 ΔcomK ΔcomS) were constructed. Deletion of the native comK gene resulted in a 50-fold decrease in transformation efficiency. Obviously, comK expressed by PcomK remained necessary for effective transformation. Deletion of both comK and comS (REG35) drastically decreased the transformation rate (Figure 5). Although, comS seemed to be less important, deletion of the original comS in REG19 reduced the number of REG32 transformants by four-fold compared to REG19 (Figure 5). Since, natural expression of comS depends on the ComP-ComA signal transduction system, the comQXPA genes were deleted in REG19 (REG37). Surprisingly, deletion of the comQXPA slightly increased the transformation efficiency of REG37 as compared to REG19 (Figure 5). Apparently, the expression of original comK and comS are necessary for a high transformation efficiency of REG19.
Figure 5.
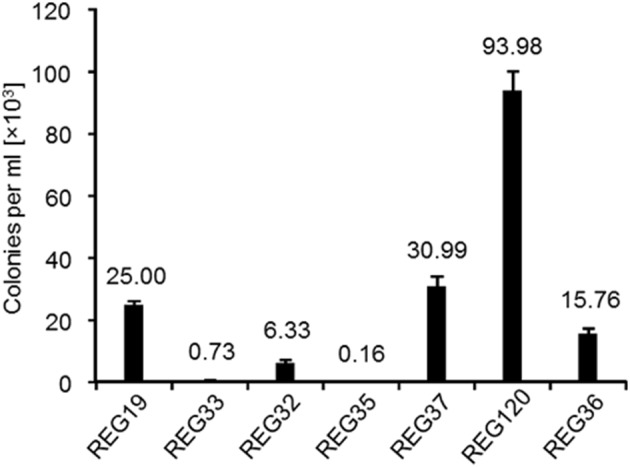
Deletion of the competence development genes in REG19 (control) constructing REG32 (ΔcomS), REG33 (ΔcomK), REG35 (ΔcomK ΔcomS), REG36 (Δspo0A), REG37 (ΔcomQXPA), REG120 (ΔdegU), and their effect on transformation efficiency. All strains were transformed in LB with 100 ng of pWAL275 dimers after induction with mannitol. Error bars represent standard deviation from the mean value between triplicate experiments.
The effect of global regulators DegU and Spo0A on transformation efficiency of REG19
At the onset of competence development, when ComK concentration is still insufficient for activation of its own gene, the non-phosphorylated response regulator DegU facilitates binding of ComK to PcomK (Hamoen et al., 2000). In contrast, phosphorylated DegU leads away from competence and activates swarming motility, biofilm formation and protease secretion (Kobayashi, 2007; Verhamme et al., 2007). To investigate the necessity of DegU for competence development in REG19, degU was deleted in REG19 (REG120). In REG120, the transformation efficiency was nearly quadrupled in comparison to REG19 (Figure 5). Another important factor for competence is Spo0A (Sadaie and Kada, 1983; Albano et al., 1987). Phosphorylated Spo0A limits the intracellular level of AbrB, which represses the transcription of comK. Mirouze et al. (2012) showed that a low level of Spo0A~P induces comK expression, whereas its high level reduces the comK expression. In REG36 (REG19 Δspo0A), the transformation efficiency was slightly reduced, from 25 × 103 colonies per ml in REG19 to 15 × 103 colonies per ml in REG36 (Figure 5). Obviously, the ectopic expression of comK and comS can compensate the need of Spo0A~P in the comK expression by its own promoter, while DegU is not further necessary for the onset of comK expression.
Deletion of chromosomal genes in REG19 using Gibson assembly
A usual way for deletion of target chromosomal DNA is its replacement with an antibiotic resistance gene via the homologous recombination. Since assembling a deletion cassette on a plasmid is time-consuming, such deletions could be accomplished after direct fusion of the amplified fragments followed by transformation of the host strain with high transformation efficiency. To demonstrate the feasibility of this method in REG19, the amyE gene was deleted using two types of ′amyE-spcR-amyE′ cassettes, which were assembled by Gibson assembly. In the first type, the end of the deletion cassettes could not overlap each other (No Repeated Sequences at the Outer Ends; NRSOE), whereas in the second type, each cassette contained 20 bp overlapping ends enabling the formation of concatemers or circular DNA fragments (Repeated Sequences at the Outer Ends; RSOE). After transformation of REG19 with NRSOE, about 200 spectinomycin resistant colonies per ml were obtained (Figure 6). All spectinomycin resistant colonies were amylase-negative on starch agar plates. Using RSOE fragments, there was a 25-fold increase in the number of spectinomycin resistant colonies compared to NRSOE fragments. This increased efficiency could be due to protection of the cassettes ends from nuclease digestion. Likewise, these overlapping ends allowed the assembly of cassettes to form larger linear fragments. This improves the uptake of single-stranded DNA which is long enough for efficient recombination. Despite the higher transformation efficiency, only 80% of the spectinomycin resistant transformants of NRSOE were amylase negative. This could be caused by circular cassettes integrated into the chromosome via single cross-over. Overall, high transformation rate of REG19 facilitates deletion of the target gene using assembled deletion cassettes without construction of plasmids.
Figure 6.
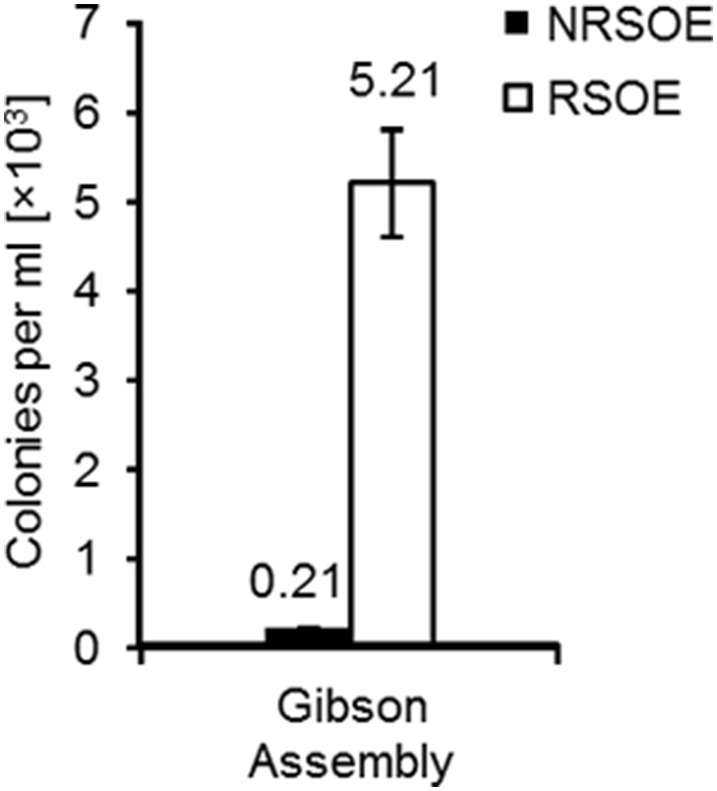
Deletion of amyE in REG19 using linear or circular DNA fragments created by Gibson assembly. The amyE deletion cassette consisted of two 700 bp flanking fragments homolog to the up-and downstream of amyE and a spectinomycin resistance gene in between. While NRSOE represents the single deletion cassettes generated by Gibson assembly, RSOE deletion cassettes were the mixture of long concatemer or circular deletion cassettes due to the overlapping regions at the both ends of each deletion cassette. After transformation of REG19, deletion of the amyE was verified in the spectinomycin resistant colonies by cultivation on starch agar. Error bars represent standard deviation from the mean value between triplicate experiments.
Discussion
In this study, we constructed a super competent B. subtilis strain (strain REG19) whose transformability is widely independent of the natural competence development. The natural competence development in B. subtilis is generally categorized into three types of regulations, namely (i) nutritional, (ii) cell-type, and (iii) growth stage specific (Dubnau, 1991b). In rich media, nutritional regulation is exerted by CodY repressing the promoters of comK and comS in the presence of branched-chain amino acids and high GTP pool (Serror and Sonenshein, 1996; Figure 1). Here, by insertion of comK under control of PmtlA, the B. subtilis cells became transformable in LB (strain REG6; Figure 2). Expressing comK with its natural promoter (van Sinderen and Venema, 1994), Pspac (Nijland et al., 2010), or PxylA (Zhang and Zhang, 2011) similarly enabled the transformation of B. subtilis in rich media. The cell-type specific regulation or comK heterogenous expression in different cells is caused by autoregulatory loop of comK (Maamar and Dubnau, 2005). Surprisingly, deletion of original comK (REG33) or comS (REG32) in the REG19 strain had a severe impact on transformation (Figure 5). This suggests that the amount of expressed ComK and ComS by PmtlA is insufficient to pass the ComK threshold stimulating competence development. Only when PmtlA was exchanged with stronger PlicB (REG89), the cells were no longer dependent on original comK (compare REG89 with REG159 in Supplementary Materials; Figure S1).
The growth stage-regulation mainly depends on the quorum sensing in a minimal medium with high cell density. To eliminate this regulation, we also overexpressed comS in addition to comK which dramatically increased the transformation efficiency (compare strains REG6 and REG19 in Figure 3). This suggests that the expression of comS remained a barrier for the full transformability of the cells in most of developed systems so far. Although, the ectopic expression of comK/comS increased the transformability and rendered competence free of its natural development process, the natural competence regulators remained effective on the REG19 transformability. Notably, the phenotypes of the mutants were in some cases different from the same deletions in the wild type strain. For instance, a slight reduction of the transformation efficiency was observed in REG36 (REG19 Δspo0A) compared to REG19 (Figure 5). In a low concentration of Spo0A(~P), the comK expression is increased by Spo0A(~P) binding to its high affinity binding sites at the comK promoter, where it antagonizes the binding of Rok. On the other hand, high Spo0A(~P) concentration represses comK expression when Spo0A(~P) binds to its low-affinity binding sites overlapping the PcomK core elements (Mirouze et al., 2012; Figure 1). Apart from its direct influence on PcomK, Spo0A(~P) downregulates the AbrB expression, thereby it renders PcomK free of AbrB repression (Smits et al., 2007). Besides, high level of the AbrB concentration represses the expression of sigH which is important for the expression of phr genes. In the latter case, the DNA binding ability of ComA(~P) is influenced by the Spo0A deletion (Auchtung et al., 2006). Surprisingly, deletion of comQXPA in REG19 (strain REG37; Figure 5) had a slightly positive effect on transformability. In the wild type strain, this deletion could result in reduction or loss of transformability (Hahn and Dubnau, 1991); however, ComA(~P) regulates many genes other than comS (Ogura et al., 2001). Thus, the observed positive effect of the comQXPA deletion could be an indirect effect. The same argument might be true for the positive effect of degU deletion. The high level of transformability in REG120 could be explained by the possibility that the degU deletion eliminates routes of differentiation contrary to competence, such as flagellar based motility (Amati et al., 2004), biofilm formation as well as protease secretion (Msadek et al., 1990; Dahl et al., 1992; Kovács and Kuipers, 2013). This deletion supports the conclusion that ComK and ComS synthesized via PmtlA compensate dephosphorylated DegU.
In conclusion, a super competent B. subtilis strain was constructed in this study by induction of comK and comS under control of PmtlA. The high transformability of this strain allows fast strain engineering using PCR fragments assembled by Gibson assembly kit. This transformation system is being successfully used for genome reduction of B. subtilis (Altenbuchner, unpublished data).
Author contributions
RR designed and performed the experiments and also analyzed the data. KM constructed some of the plasmids and strains used in this study. Also, KM was involved in data analysis and manuscript preparation. JA directed and coordinated the project. The final paper was written by RR and KM which was later revised and corrected by JA. All authors approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Jörg Stülke for his helpful suggestions during manuscript preparation. We would like to thank Lydia Warth for construction of the pWAL275 plasmid. We are also grateful to Annette Schneck for her technical assistance.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01431
References
- Albano M., Hahn J., Dubnau D. (1987). Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169, 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbuchner J., Viell P., Pelletier I. (1992). Positive selection vectors based on palindromic DNA sequences. Methods Enzymol. 216, 457–466. 10.1016/0076-6879(92)16042-I [DOI] [PubMed] [Google Scholar]
- Amati G., Bisicchia P., Galizzi A. (2004). DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J. Bacteriol. 186, 6003–6014. 10.1128/JB.186.18.6003-6014.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. (1961). Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung J. M., Lee C. A., Grossman A. D. (2006). Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 188, 5273–5285. 10.1128/JB.00300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka R. M., Hahn J., Albano M., Draskovic I., Persuh M., Cui X., et al. (2002). Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43, 1331–1345. 10.1046/j.1365-2958.2002.02833.x [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951). Studies on lysogenesis I.: the mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. (1991). Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64, 545–552. 10.1016/0092-8674(91)90238-T [DOI] [PubMed] [Google Scholar]
- Chung C. T., Niemela S. L., Miller R. H. (1989). One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 86, 2172–2175. 10.1073/pnas.86.7.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comella N., Grossman A. D. (2005). Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 57, 1159–1174. 10.1111/j.1365-2958.2005.04749.x [DOI] [PubMed] [Google Scholar]
- Dahl M. K., Msadek T., Kunst F., Rapoport G. (1992). The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267, 14509–14514. [PubMed] [Google Scholar]
- Dubnau D., Provvedi R. (2000). Internalizing DNA. Res. Microbiol. 151, 475–480. 10.1016/S0923-2508(00)00166-2 [DOI] [PubMed] [Google Scholar]
- Dubnau D. (1991a). Genetic competence in Bacillus subtilis. Microbiol. Rev. 55, 395–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. (1991b). The regulation of genetic competence in Bacillus subtilis. Mol. Microbiol. 5, 11–18. 10.1111/j.1365-2958.1991.tb01820.x [DOI] [PubMed] [Google Scholar]
- Duitman E. H., Wyczawski D., Boven L. G., Venema G., Kuipers O. P., Hamoen L. W. (2007). Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Appl. Environ. Microbiol. 73, 3490–3496. 10.1128/AEM.02751-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A.-M., Lehrach H., Poustka A., Murray N. (1983). Lambda replacement vectors carrying polylinker sequences. J. Mol. Biol. 170, 827–842. 10.1016/S0022-2836(83)80190-9 [DOI] [PubMed] [Google Scholar]
- Gamba P., Jonker M. J., Hamoen L. W. (2015). A novel feedback loop that controls bimodal expression of genetic competence. PLoS Genet. 11:e1005047. 10.1371/journal.pgen.1005047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pastor J. E., Hobbs E. C., Losick R. (2003). Cannibalism by sporulating bacteria. Science 301, 510–513. 10.1126/science.1086462 [DOI] [PubMed] [Google Scholar]
- Hahn J., Dubnau D. (1991). Growth stage signal transduction and the requirements for srfA induction in development of competence. J. Bacteriol. 173, 7275–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Luttinger A., Dubnau D. (1996). Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol. 21, 763–775. 10.1046/j.1365-2958.1996.371407.x [DOI] [PubMed] [Google Scholar]
- Hahn J., Roggiani M., Dubnau D. (1995). The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J. Bacteriol. 177, 3601–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen L. W. (2011). Cell division blockage: but this time by a surprisingly conserved protein. Mol. Microbiol. 81, 1–3. 10.1111/j.1365-2958.2011.07693.x [DOI] [PubMed] [Google Scholar]
- Hamoen L. W., Smits W. K., de Jong A., Holsappel S., Kuipers O. P. (2002). Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30, 5517–5528. 10.1093/nar/gkf698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen L. W., van Werkhoven A. F., Venema G., Dubnau D. (2000). The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 97, 9246–9251. 10.1073/pnas.160010597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen L. W., Venema G., Kuipers O. P. (2003). Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149, 9–17. 10.1099/mic.0.26003-0 [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Cutting S. M. (1990). Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons Ltd. [Google Scholar]
- Hoa T. T., Tortosa P., Albano M., Dubnau D. (2002). Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43, 15–26. 10.1046/j.1365-2958.2002.02727.x [DOI] [PubMed] [Google Scholar]
- Inamine G. S., Dubnau D. (1995). ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J. Bacteriol. 177, 3045–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane D., Ayora S., Sweasy J. B., Graumann P. L., Alonso J. C. (2012). The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery. Crit. Rev. Biochem. Mol. Biol. 47, 531–555. 10.3109/10409238.2012.729562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. (2007). Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66, 395–409. 10.1111/j.1365-2958.2007.05923.x [DOI] [PubMed] [Google Scholar]
- Kong L., Dubnau D. (1994). Regulation of competence-specific gene expression by Mec-mediated protein-protein interaction in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 91, 5793–5797. 10.1073/pnas.91.13.5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács A. T., Kuipers O. P. (2013). Transcriptional Circuits and Phenotypic Variation. Norfolk, VA: Caister Academic Press. [Google Scholar]
- Liu L., Nakano M. M., Lee O. H., Zuber P. (1996). Plasmid-amplified comS enhances genetic competence and suppresses sinR in Bacillus subtilis. J. Bacteriol. 178, 5144–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H., Dubnau D. (2005). Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56, 615–624. 10.1111/j.1365-2958.2005.04592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R., Solomon J., Grossman A. D. (1994). Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77, 207–216. 10.1016/0092-8674(94)90313-1 [DOI] [PubMed] [Google Scholar]
- Marsh J., Erfle M., Wykes E. J. (1984). The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32, 481–485. 10.1016/0378-1119(84)90022-2 [DOI] [PubMed] [Google Scholar]
- Mirouze N., Desai Y., Raj A., Dubnau D., Hughes D. (2012). Spo0A~P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet. 8:e1002586. 10.1371/journal.pgen.1002586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motejadded H., Altenbuchner J. (2007). Integration of a lipase gene into the Bacillus subtilis chromosome: recombinant strains without antibiotic resistance marker. Iran. J. Biotech. 5, 105–109. [Google Scholar]
- Msadek T., Kunst F., Henner D., Klier A., Rapoport G., Dedonder R. (1990). Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172, 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland R., Burgess J. G., Errington J., Veening J.-W., Redfield R. J. (2010). Transformation of Environmental Bacillus subtilis Isolates by Transiently Inducing Genetic Competence. PLoS ONE 5:e9724. 10.1371/journal.pone.0009724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara T., Freese E. (1975). Motility of Bacillus subtilis during growth and sporulation. J. Bacteriol. 123, 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Yamaguchi H., Yoshida K., Fujita Y., Tanak T. (2001). DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29, 3804–3813. 10.1093/nar/29.18.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Hilbert D. W. (2004). Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7, 579–586. 10.1016/j.mib.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Prepiak P., Defrancesco M., Spadavecchia S., Mirouze N., Albano M., Persuh M., et al. (2011). MecA dampens transitions to spore, biofilm exopolysaccharide and competence expression by two different mechanisms. Mol. Microbiol. 80, 1014–1030. 10.1111/j.1365-2958.2011.07627.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D. Z., Losick R. (2001). Morphological Coupling in Development: lesson form prokaryotes. Dev. Cell 1, 733–742. 10.1016/S1534-5807(01)00094-6 [DOI] [PubMed] [Google Scholar]
- Sadaie Y., Kada T. (1983). Formation of competent Bacillus subtilis cells. J. Bacteriol. 153, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Serror P., Sonenshein A. L. (1996). Interaction of CodY, a novel Bacillus subtillis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20, 843–852. 10.1111/j.1365-2958.1996.tb02522.x [DOI] [PubMed] [Google Scholar]
- Smits W. K., Eschevins C. C., Susanna K. A., Bron S., Kuipers O. P., Hamoen L. W. (2005). Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56, 604–614. 10.1111/j.1365-2958.2005.04488.x [DOI] [PubMed] [Google Scholar]
- Smits W. K., Hoa T. T., Hamoen L. W., Kuipers O. P., Dubnau D. (2007). Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein. Mol. Microbiol. 64, 368–381. 10.1111/j.1365-2958.2007.05662.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. (1958). Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U.S.A. 44, 1072–1078. 10.1073/pnas.44.10.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse S., Graumann P. L. (2007). DprA/Smf protein localizes at the DNA uptake machinery in competent Bacillus subtilis cells. BMC Microbiol. 7:105. 10.1186/1471-2180-7-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay K., Hahn J., Burghoorn J., Dubnau D. (1998). Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17, 6730–6738. 10.1093/emboj/17.22.6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay K., Hamoen L. W., Venema G., Dubnau D. (1997). Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11, 119–128. 10.1101/gad.11.1.119 [DOI] [PubMed] [Google Scholar]
- van Sinderen D., Venema G. (1994). ComK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J. Bacteriol. 176, 5762–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme D. T., Kiley T. B., Stanley-Wall N. R. (2007). DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65, 554–568. 10.1111/j.1365-2958.2007.05810.x [DOI] [PubMed] [Google Scholar]
- Vlamakis H., Aguilar C., Losick R., Kolter R. (2008). Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953. 10.1101/gad.1645008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth L., Haug I., Altenbuchner J. (2011). Characterization of the tyrosine recombinase MrpA encoded by the Streptomyces coelicolor A3(2) plasmid SCP2*. Arch. Microbiol. 193, 187–200. 10.1007/s00203-010-0662-9 [DOI] [PubMed] [Google Scholar]
- Wenzel M., Altenbuchner J. (2015). Development of a markerless gene deletion system for Bacillus subtilis based on the mannose phosphoenolpyruvate-dependent phosphotransferase system. Microbiology 161, 1942–1949. 10.1099/mic.0.000150 [DOI] [PubMed] [Google Scholar]
- Yadav T., Carrasco B., Hejna J., Suzuki Y., Takeyasu K., Alonso J. C. (2013). Bacillus subtilis DprA recruits RecA onto ssDNA and mediates annealing of complementary ssDNA strands coated by SsbB and SsbA. J. Biol. Chem. 288, 22437–22450. 10.1074/jbc.M113.478347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33, 103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- Zhang X.-Z., Zhang Y.-H. P. (2011). Simple, fast and high-efficiency transformation system for directed evolution of cellulase in Bacillus subtilis. Microb. Biotechnol. 4, 98–105. 10.1111/j.1751-7915.2010.00230.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.