Abstract
Single-chain variable antibody fragments (scFvs) are attractive candidates for targeted immunotherapy in several human diseases. In this study, a concise humanization strategy combined with an optimized production method for humanizing scFvs was successfully employed. Two antibody clones, one directed against the hemagglutinin of H5N1 influenza virus, the other against EpCAM, a cancer biomarker, were used to demonstrate the validity of the method. Heavy chain (VH) and light chain (VL) variable regions of immunoglobulin genes from mouse hybridoma cells were sequenced and subjected to the construction of mouse scFv 3-D structure. Based on in silico modeling, the humanized version of the scFv was designed via complementarity-determining region (CDR) grafting with the retention of mouse framework region (FR) residues identified by primary sequence analysis. Root-mean-square deviation (RMSD) value between mouse and humanized scFv structures was calculated to evaluate the preservation of CDR conformation. Mouse and humanized scFv genes were then constructed and expressed in Escherichia coli. Using this method, we successfully generated humanized scFvs that retained the targeting activity of their respective mouse scFv counterparts. In addition, the humanized scFvs were engineered with a C-terminal cysteine residue (hscFv-C) for site-directed conjugation for use in future targeting applications. The hscFv-C expression was extensively optimized to improve protein production yield. The protocol yielded a 20-fold increase in production of hscFv-Cs in E. coli periplasm. The strategy described in this study may be applicable in the humanization of other antibodies derived from mouse hybridoma.
Introduction
Single-chain variable antibody fragments (scFvs) have enormous potential in clinical applications. ScFv is an excellent targeting ligand for in vivo cancer imaging, as well as for mediating cell targeting in drug delivery systems. Its small structure, containing only the antigen binding site (about 30 kDa rather than 150 kDa of IgG), promotes tissue penetration and speeds up clearance time.(1–3)
There are two common strategies for generating scFvs: phage display or cloning of variable regions from mouse hybridoma.(4,5) Despite the popularity of scFv antibodies generated by phage display, obtaining high affinity scFvs from phage libraries remains a challenging task.(6) Meanwhile, mouse hybridoma is the predominant source of monoclonal antibodies (MAbs) that are well characterized with high affinity against different targets. Thus far the available therapeutic scFvs are constructed primarily from mouse hybridoma.(7–9)
Generally, scFvs are engineered to contain an antigen-binding site by cloning heavy and light chain variable region genes (VH and VL) from hybridoma cells that secrete MAbs. The VH and VL regions are linked with a flexible polypeptide linker, (Gly4Ser)3.(5) For targeting applications, scFvs can also be engineered by adding a free cysteine at the carboxyl end of the structure.(10) Applicability of cysteine-tagged scFvs for site-directed conjugation has been reported, specifically, in site-specific covalent radioactive labeling and site-specific conjugation to lipids in liposomes.(11,12)
Engineering of humanized scFv from mouse scFv is essential for the generation of therapeutic agents. A variety of antibody humanization techniques to reduce human anti-mouse antibody (HAMA) responses has been developed.(13–15) The standard method involves grafting mouse complementarity-determining regions (CDRs) onto human framework regions (FRs). The critical objective is to prevent loss of antigen-binding affinity due to loss of original CDR conformations after CDR grafting.(16,17) Several factors play a role in preventing loss of affinity, including proper selection of human template, compatibility between mouse CDRs and human FRs, and retention or back mutation of mouse FR residues at positions that maintain original CDR conformation.(18,19) Each back mutation can be individually defined by computer-assisted molecular modeling and sometimes requires trials of many different variants of the CDR-grafted antibodies to identify back mutations.(20,21) In some cases, back mutations at well-defined positions are counterproductive. To correct this problem, a simple and efficient humanization strategy combined with an analytical method to predict the preservation of original CDR conformation could lead to more successful antibody humanization.
The present study demonstrates a simple, but effective humanization method for the production of humanized scFvs from mouse hybridomas. The method is based on generic CDR grafting, with some modifications. Key mouse FR residues, identified by primary sequence analysis, are retained onto FRs of the human antibody to prevent affinity loss. Analysis of root-mean-square deviation (RMSD) between mouse and humanized scFv structures provides guidance in the identification and selection of the humanized sequences that retain the original CDR conformation. This process makes the humanization outcome more predictable and therefore more successful.
Materials and Methods
Cell lines
Colorectal cancer cell line HT-29 was cultured in McCoy's 5A modified medium (Gibco, Carlsbad, CA), supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 U/mL penicillin-streptomycin. Embryonic kidney cell line HEK-293T was cultured in RPMI (Gibco), supplemented with 10% fetal bovine serum and 100 U/mL penicillin-streptomycin. All cells were maintained at 37°C in a 5% CO2 atmosphere.
Amplification of antibody variable region genes
The variable region of heavy chain (VH) and variable region of light chain (VL) of immunoglobulin (Ig) sequences were obtained from two hybridoma clones. One clone secreting IgG2a MAb was directed against EpCAM protein (clone 2H10) and the other secreting IgG2a MAb directed against hemagglutinins (HA) of the H5N1 virus (clone 5E11). The mouse hybridoma clones were established in our laboratory. Total RNA was extracted from 1 × 106 hybridoma cells using RNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's instructions. The cDNA of VH and VL were synthesized using OneStep RT-PCR (Qiagen). Primers used for cDNA amplification of VH and VL from two antibodies were independently selected. Primers used for amplification of 2H10 VH cDNA were MH1 and IgG2, in accordance with Wang and colleagues.(22) Primers used for amplification of 2H10 VL cDNA were Vk4 and Ck, in accordance with Coloma and colleagues.(23) Consistent with Larrick and colleagues (23), primers used for amplification of 5E11 VH cDNA were VH1 and CH1. Primers used for amplification of 5E11 VL cDNA were Vk1 and Ck, in accordance with Coloma and colleagues(23). All primers were synthesized by Bio Basic (Markham, Canada; Table 1). VH and VL genes were then sequenced and CDR and FR positions were determined using the Igblast Kabat program (www.ncbi.nlm.nih.gov/igblast). CDRH:3 and CDRL:3 were predicted according to Kabat definition based on sequence variability, which is the most commonly used method for CDR3 prediction (www.bioinf.org.uk/abs/#cdrdef). Assignment of Kabat numbering to VH and VL genes was performed using Kabat sequence analysis tools (www.bioinf.org.uk/abs/abnum).(24) An automated program (www.bioinf.org.uk/abs/chothia.html) was used for determination of Chothia canonical structural class of CDRs. CDR-H3, which had the most diverse CDR structure, was not analyzable by the program.
Table 1.
Primer Sequences Used for Amplification of VH And VL Genes of Mouse MAbs
| Primer | Sequence 5′ to 3′ |
|---|---|
| 2H10 VH and VL genes | |
| MH1 | SARGTNMAGCTGSAGSAGTC |
| IgG2 | CTTGACCAGGCATCCTAGAGTCA |
| Vk4 | CACCATGKCCCCWRCTCAGYTYCTTGT |
| Ck | GGGGTCGACACTGGATGGTGGGAAGATGGA |
| 5E11 VH and VL genes | |
| VH1 | GGGGATATCCACCATGGRATGSAGCTGKGTMATSCTCTT |
| CH1 | GGGGCTAGCYCTCCACACACAGGRRCCAGTGGATAGAC |
| Vk1 | GGGGATATCCACCATGGAGACAGACACACTCCTGCTAT |
| Ck | GGGGTCGACACTGGATGGTGGGAAGATGGA |
Degenerate nucleotides: K = G or T; R = A or G; S = G or C; W = A or T; Y = C or T; M = A or C; N = A or C (or T or G).
Design of humanized single-chain antibody
The humanization design process involved CDR grafting and used three-dimensional (3-D) structure information to guide the process. To generate the humanized scFv gene, six CDRs of mouse VH and VL were grafted onto selected human FRs showing the highest amino acid sequence identity to the FRs of mouse VH and VL. The human sequences used in the humanization of the two scFvs were different. The human immunoglobulin germline sequence was used as the selected human FRs for 2H10, while the human immunoglobulin consensus sequence was used as the selected human FRs for 5E11. Human immunoglobulin germline sequences showing the highest amino acid sequence similarity in FRs between human and 2H10 VH and VL were identified independently using IMGT/V-QUEST (www.imgt.org/IMGT_vquest/vquest?livret=0&Option=humanIg).
Human immunoglobulin consensus sequences that showed the highest amino acid sequence similarity in FRs between human and 5E11 VH and VL were identified independently from human antibody sequences from the Kabat database (http://people.cryst.bbk.ac.uk/∼ubcg07s/cons/cons.html). Amino acid residues in FRs of mouse VH and VL that differed from human FRs were substituted with human residues, while preserving mouse residues at positions known as Vernier zone residues and chain-packing residues.
In order to study the importance of FR residues that could influence CDR conformation, 3-D structural models of mouse and humanized scFvs were constructed. Three-dimensional structural models of mouse 2H10scFv (m2H10scFv) and mouse 5E11scFv (m5E11scFv) were constructed using computer-assisted molecular modeling, based on VH and VL with highly identical amino acid sequences and 3-D structures. The Protein Data Bank (PDB) was searched for antibody sequences with a close sequence identity to VH and VL of 2H10 and 5E11. Two separate BLASTp searches were performed for VH and VL. The amino acid sequence of antibody ID 2I9L chain B and 3CFB chain L showed 88% and 97% identity with VH and VL of 2H10, respectively. Similarly, the amino acid sequence of antibody ID 3S35 chain H and 4FFZ chain L showed 78% and 94% identity with VH and VL of 5E11, respectively. This close sequence identity facilitated construction of a 3-D model of mouse scFvs using the software package SwissPdb Viewer (http://spdbv.vital-it.ch). Newly built structures were subjected to refined 3-D modeling using SWISS MODEL program (www.expasy.ch/swissmod/SWISS-MODEL.html). All structures were solvated using TIP5P water model, with 2000 steps of minimization subsequently performed. The structure was equilibrated for 0.1 ns before a 3 ns production run at 300 K using the SANDER module of AMBER9. Equilibrium structure of mouse scFv was obtained by averaging molecular dynamics (MD) simulation results of the antibody between 2 and 3 ns. For the 3-D structural model of humanized scFv, a single residue was manually changed from the mouse scFv 3-D structure using the Swiss PDB Viewer program. The created model was then subjected to energy minimization under the same conditions used for mouse scFv. The resulting models were viewed and analyzed using the Swiss PDB Viewer and PyMOL (www.pymol.org).
Residues that differed between mouse and selected human FRs were determined individually for compatibility of substitutional changes, as guided by the 3-D structural model of the mouse scFv. A root-mean-square deviation (RMSD) value of the two superimposed scFv structures was calculated on alpha carbon atom using the Swiss PDB Viewer program.
Construction, expression, and purification of scFvs
The genes encoding the mouse scFv and humanized scFv were synthesized in VH-linker-VL format with standard 15 amino acid linker (Gly4Ser)3. In cases of cysteine-tagged humanized scFv (hscFv-C), genes were synthesized in VH-(Gly4Ser)3-VL-6xHis-(GS)2-Cys format. All scFv genes were codon optimized and synthesized by GenScript USA (Piscataway, NJ). Synthetic genes were inserted into a pET26b+ vector containing a pelB promoter for controlling periplasmic protein expression (Novagen, Madison, WI). For scFv expression, E. coli RosettaBlue (DE3) cells were transformed with recombinant plasmids (Novagen). Transformed E. coli Rosettablue (DE3) cells were cultured at 37°C in LB medium with 50 μg/mL kanamycin and 12.5 μg/mL tetracycline until OD600 reached 0.6, then induced with 1 mM IPTG at 20°C for 18 h. For hscFv-C expression, condition optimization, such as decreasing the temperature, varying amounts of IPTG, and adding different concentrations of glycine (1–3% W/V), Triton X-100 (1–3% V/V), or 0.4 M sucrose to the culture medium after induction were tested. Soluble scFvs were extracted from E. coli periplasm using TSE buffer (300 mM Tris-HCl [pH 8.0], 20% sucrose, and 1 mM EDTA). The periplasmic fraction was purified using HisTrap FF affinity chromatography column (GE Healthcare, Uppsala, Sweden), in accordance with the manufacturer's instructions. The purity of purified scFvs was determined by 12% SDS-PAGE.
Expression of recombinant hemagglutinin in baculovirus expression system
The recombinant hemagglutinin (rHA) of influenza A H5N1 was expressed in Spodoptera frugiperda (sf9) using the baculovirus expression system. Briefly, the full length of HA gene of H5N1 virus was amplified from influenza A/chicken/Nakorn-Pathom/Thailand/cu-k2/2004(H5N1) using QIAamp Viral RNA Mini Kit (Qiagen). The amplified gene was then cloned into pFastBacHtb, a donor plasmid (Invitrogen, Carlsbad, CA). The recombinant pFastBacHtb was then transformed into E. coli DH10BAC, a baculovirus shuttle vector (Invitrogen), to create recombinant bacmid. Then, sf9 cells were transfected with recombinant bacmid to produce recombinant baculovirus. For recombinant HA protein expression, sf9 cells were infected with recombinant baculovirus at multiplicity of infection (MOI) of 1, for 96 h. The sf9 expressing HA cells were harvested and subjected to either crude rHA extraction or rHA purification. The resulting rHA was then used as antigen in ELISA experiments for testing binding activity of 5E11 antibodies.
Analysis of antigen binding specificity of scFvs by competitive ELISA
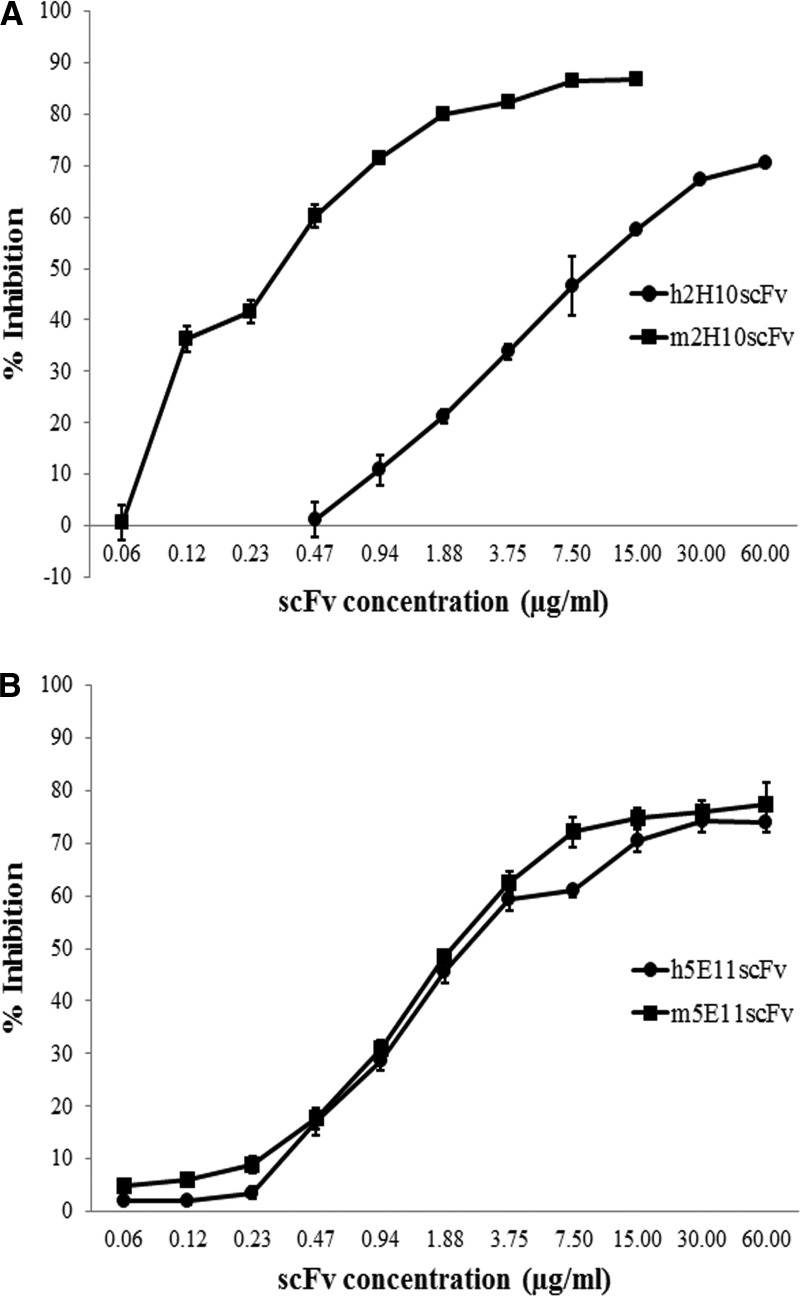
Competitive ELISA was performed to confirm that scFvs bind to the same epitope as the parental antibodies. For humanized 2H10scFv (h2H10scFv), HT29 cells (25,000 cells/well) were seeded in 96-well cell culture plate and allowed to attach to the well surface. After growing in media for 24 h, cells were fixed with 1:1 acetone-methanol and blocked at 37°C for 1 h with 2% skimmed milk. Cells were then washed with PBST (PBS with 0.05% Tween-20). Different concentrations of purified scFvs were added to the wells in the presence of 2H10 MAb (0.4 μM) and allowed to compete for 1 h at 37°C. After incubation, cells were washed with PBST and bound 2H10 MAb was detected using HRP-conjugated anti-mouse IgG antibody (KPL, Gaithersburg, MD) and TMB substrate (KPL). The plate was then read in an ELISA reader at 450/630 nm.
For humanized 5E11scFv (h5E11scFv), the ELISA plate was coated at 4°C overnight with 1:200 crude extract of rHA and then blocked with 3% BSA in PBST at 37°C for 1 h. Different concentrations of scFvs were added to the antigen-coated ELISA plate in the presence of a 1:1500 dilution of biotin-conjugated 5E11 MAb and were allowed to compete for 1 h at 37°C. The plate was washed with PBST and bound 5E11 MAb was detected using HRP-conjugated streptavidin (Thermo Scientific, Waltham, MA) and TMB substrate (KPL). The plate was then read in an ELISA reader at 450/630 nm. Percent inhibition was calculated using [1 – (A2/A1)] × 100 = percent inhibition for any given concentration of inhibitor, with A1 = absorbance in the absence of inhibitor (maximum absorbance) and A2 = absorbance at any given concentration.
Cell targeting ability of humanized scFv by immunofluorescence staining
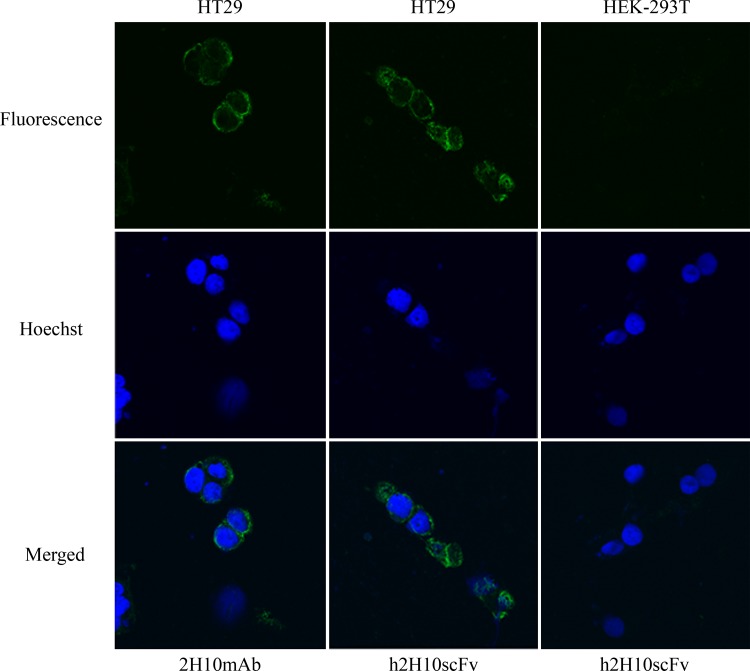
HT29 (EpCAM positive cell line) or HEK293T cells (EpCAM negative cell line) at 60,000 cells/well were seeded in an 8-well chamber slide and allowed to attach to the well surface. After growing in media for 24 h, cells were washed with PBS and then blocked with 1% BSA and 10% goat serum in PBS for 30 min at room temperature. After blocking, cells were incubated with 2H10 MAb or scFv for 1 h on ice. In the case of scFv, cells were additionally incubated with mouse anti-histidine (Invitrogen). Bound antibodies on cells were detected by FITC-conjugated goat anti-mouse IgG (Invitrogen) containing Hoechst 33342 (Thermo Scientific). After being washed with PBS, slides were mounted with 50% glycerol and covered with a cover slip. Immunofluorescence of cells was visualized with confocal laser scanning (Zeiss LSM 510 META, Carl Zeiss, Jena, Germany).
Antibody affinity measurement by ELISA
Antibody affinity was determined using the described method with some modifications.(25) Affinity of 2H10 MAb and scFvs was measured using cell-based indirect ELISA. Different concentrations of HT29 cells (1.25, 2.5, 5 of 10 × 105 cells) were seeded in a 96-well cell culture plate and allowed to attach to the well surface. After growing in media for 24 h, cells were fixed and blocked as described in the competitive ELISA section above. Cells were then incubated with serial dilutions of 2H10 MAbs 37°C for 1 h. All wells were then washed, followed by the addition of HRP-conjugated goat anti-mouse IgG and further incubation at 37°C for 30 min. In the case of 2H10scFvs, cells were incubated with mouse anti-histidine at 37°C for 30 min and washed before the addition of HRP-conjugated goat anti-mouse IgG and further incubation at 37°C for 30 min. The color was developed with TMB solution (KPL), and the plate was read in an ELISA reader at 450/630 nm. Affinity constants (Kaff) of antibodies for EpCAM found on the surface of HT29 cells were calculated using the equation: Kaff = (n – 1)/2(n[Ab’]t2[Ab]t), where n = [Ag]/[Ag’]. Briefly, [Ag] and [Ag’] represent the amount of HT29 cells or rHA; [Ab’]t and [Ab]t represent measurable total antibody concentrations at half the maximum OD (OD-50) for plates coated with [Ag’] and [Ag], respectively.
In the case of 5E11 MAb, different concentrations of purified rHA (0.25, 0.5, 1, 2 μg/mL) were coated onto the ELISA plate and incubated with serial dilutions of MAbs at 37°C for 1 h. The plate was then washed and incubated with HRP-conjugated goat anti-mouse IgG at 37°C for 30 min. In the case of 5E11scFvs, protein L-conjugated HRP was added to each well and further incubated at 37°C for 30 min. The color was developed with TMB solution (KPL) and the plate was read in an ELISA reader at 450/630 nm. Affinity constants of antibodies for rHA were calculated using the same equation described above.
Statistical analysis
All ELISA experiments were performed in triplicate, with data expressed as mean ± SD.
Results
Design of humanized single-chain antibody
Hybridoma cells secreting IgG2a MAbs directed against EpCAM (clone 2H10) and hemagglutinins (HA) of H5N1 virus (clone 5E11) were used as starting materials. Genes encoding VH and VL were amplified by RT-PCR using a set of degenerate primers. VH and VL genes were then constructed into mouse scFv sequences. Three-dimensional structural models of mouse scFvs were simultaneously constructed using homology modeling.
Humanization of m2H10scFv and m5E11scFv was accomplished by CDR grafting. The strategy employed in the selection of human sequences that showed the highest sequence similarity in FRs between mouse and human, as well as similarity in the canonical structural class of CDRs. A set of six CDRs from the VH and VL regions were then grafted onto the corresponding regions of the selected human sequences. In this study, the selected human sequences used in the humanization of the two scFvs were different. To design the humanized 2H10scFv (h2H10scFv) sequence, the human immunoglobulin germline, the V region from IGHV1-46*03 group, and the J region from IGHJ4*03 group were selected as human acceptor FRs, as they showed 75.8% amino acid identity to FRs of 2H10 VH. For VL, the human immunoglobulin germline, the V region from IGKV2-28*03 group, and the J region from IGKJ2*01 group were selected as human acceptor FRs, as they showed 88.7% amino acid identity to the FRs of 2H10 VL.
In the case of h5E11scFv, the FR of human immunoglobulin consensus sequences, VH subgroup I and VLκ subgroup IV were used as acceptor FRs, as they showed 79% and 81% amino acid identity to the FRs of 5E11 VH and VL, respectively. Alignments of VH and VL sequences of 2H10 and 5E11 with their selected human sequences are shown in Figure 1. In addition, the canonical structural class of CDRs between mouse and human sequence was matched in order to determine compatibility for CDR grafting. The Chothia canonical structural classes of 2H10 CDRs were H1-1, H2-2, L1-4, L2-1, and L3-1. For 5E11, the classes were H1-1, H2-2, L2-1, and L3-1, whereas the L1 class was undefined by the database. Four CDRs of the selected human sequence belonged to the same canonical class as 2H10 (H1-1, L1-4, L2-1, and L3-1). Similarly, three CDRs of the selected human sequence belonged to the same canonical class as 5E11 (H1-1, L2-1, and L3-1).
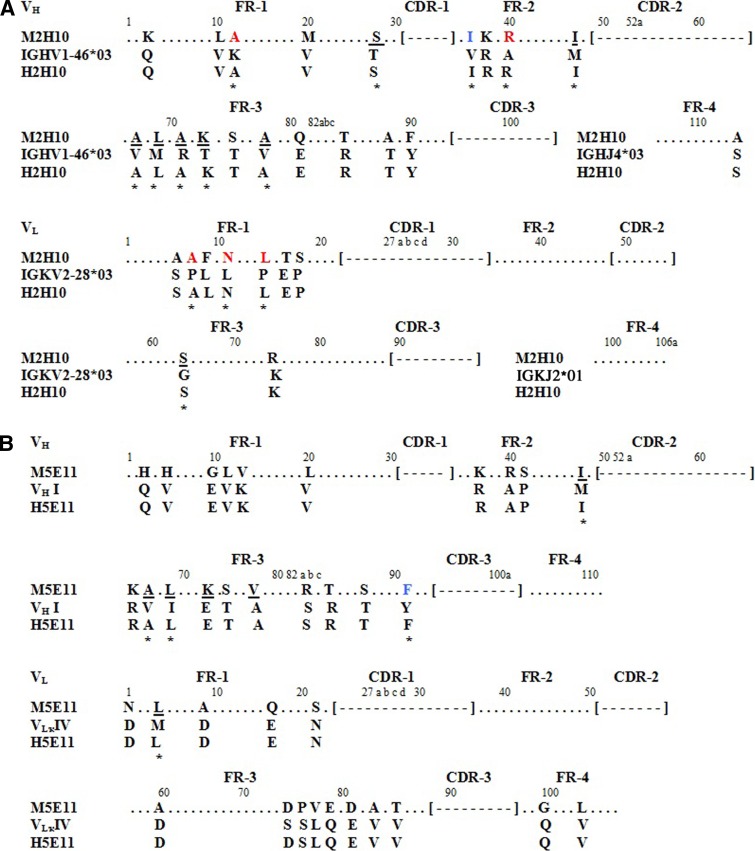
FIG. 1.
Sequence alignment of variable regions of mouse antibodies, selected human sequences, and humanized antibodies. (A) Alignment of amino acid sequences of variable regions of 2H10 with human immunoglobulin germline sequence and humanized 2H10 sequence. (B) Alignment of amino acid sequences of variable regions of 5E11 with human immunoglobulin consensus sequence and humanized 5E11 sequence. Amino acid residues are numbered according to the convention of Kabat and colleagues.(24) Identical amino acid residues are marked by dots (●). CDR residues, according to Kabat's definition, are indicated in brackets. Vernier zone residues are underlined. Chain-packing residues are blue-colored. Residues that differ in hydrophilic and hydrophobic properties are red-colored. Mouse FR residues retained in humanized antibody are indicated by asterisks (*).
Vernier zone residues have been known to critically affect antigen binding by having direct contact with the antigen or by affecting the CDR loop.(18,26) Similarly, chain-packing residues are residues buried at the VH/VL interface that affect conformation of the antigen binding pocket.(27) Because these residues may strongly affect the structure of CDRs and antibody affinity, the conventional humanization procedure suggests that these mouse residues should be retained (back mutation) at these positions.(28) In this study, we focused on the preservation of the mouse Vernier zone and chain packing residues onto FRs of the human antibody by primary sequence analysis. Sequence comparison between 2H10 and selected human FRs showed that twenty and nine amino acid residues were different in VH and VL, respectively. With a focus on Vernier zone residues, seven residues (SerH:28, IleH:48, AlaH:67, LeuH:69, AlaH:71, LysH:73, and AlaH:78) for VH, and one residue (SerL:64) for VL were different between sequences. At the same time, one of the chain-packing residues for VH (IleH:37) also differed from the human counterpart, with no such difference in the residues of VL. Accordingly, these mouse residues were retained in the h2H10scFv FRs (Fig. 1).
In the case of 5E11, sequence comparison between 5E11 and selected human FRs at the Vernier zone showed five residues (IleH:48, AlaH:67, LeuH:69, LysH:73, and ValH:78) in VH, and one residue (LeuL:4) in VL to be different between sequences. PheH:91 was the only chain-packing residue in VH that differed from the human counterpart. As such, these mouse residues were retained in the h5E11scFv FRs (Fig. 1).
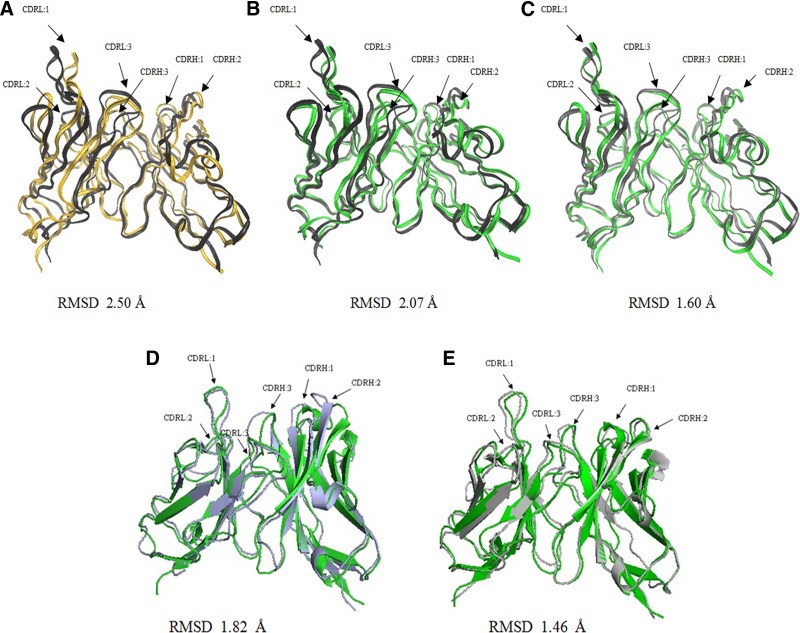
Designed humanized scFv sequences were subjected to construction of 3-D structure by manually changing a single residue from constructed mouse scFv 3-D structures. To evaluate similarity between the 3-D structures, humanized scFv was superimposed onto mouse scFv to determine RMSD value. Accordingly, h2H10scFv was superimposed onto m2H10scFv. A comparison of RMSD values before and after back mutation of the mouse Vernier zone and chain-packing residues in human FRs revealed that the RMSD of h2H10scFv reduced from 2.50 Å to 2.07 Å (Fig. 2). In order to increase similarity between h2H10scFv and its mouse scFv counterpart, we re-evaluated mouse residues for back mutation from remaining non-identical residues in VH and VL. Based on the fact that changing hydrophobic-hydrophilic interactions have a strong impact on protein folding, we focused on back mutation of remaining non-identical residues at positions that differed in hydrophilic and hydrophobic properties. Sequence comparison between 2H10 and selected human FRs showed that five amino acids were different in terms of hydrophilic and hydrophobic properties, as determined by hydrophobicity index (Fig. 1).(29,30) So, we back-mutated all of these mouse residues onto human FRs. The 3-D structure of h2H10scFv, with additional back mutation, was superimposed onto m2H10scFv to determine the impact of back-mutated residues on CDR conformation. Results showed that the sets of back mutation that consisted of LysH:12 to Ala and AlaH:40 to Arg in VH, and ProL:8 to Ala, LeuL:11 to Asn, and ProL:15 to LeuL in VL progressively reduced RMSD from 2.07 Å to 1.60 Å (Fig. 2). As a result, these mouse residues were also retained in the h2H10scFv FRs. The back mutation of mouse residues, as described above, allowed the engineered h2H10scFv sequence to become close in identity to the human sequence. The amino acid sequence identity to the selected human FR sequence of VH was 87.3% and 93.5% amino acid sequence identity to the selected human FRs sequence of VL.
FIG. 2.
Overall superimposed structures of mouse scFv and humanized scFv. (A) Structure of h2H10scFv before back mutation (orange) is superimposed onto m2H10scFv (gray). (B) Structure of h2H10scFv with back mutation of mouse residues (green) superimposed onto m2H10scFv (gray). (C) Structure of h2H10scFv with additional back mutations at positions that are different in hydrophobic and hydrophilic properties (green) superimposed onto m2H10scFv (gray). (D) Structure of h5E11scFv before back mutation (green) superimposed onto m5E11scFv (gray). (E) Structure of h5E11scFv with back mutation of mouse residues (green) superimposed onto m5E11scFv (gray).
Similarly, h5E11scFv was subsequently evaluated for RMSD value by superimposing its 3-D structure onto the m5E11scFv structure. Results showed that the RMSD of h5E11scFv reduced from 1.82 Å to 1.46 Å, compared with RMSD value before back mutation (Fig. 2).
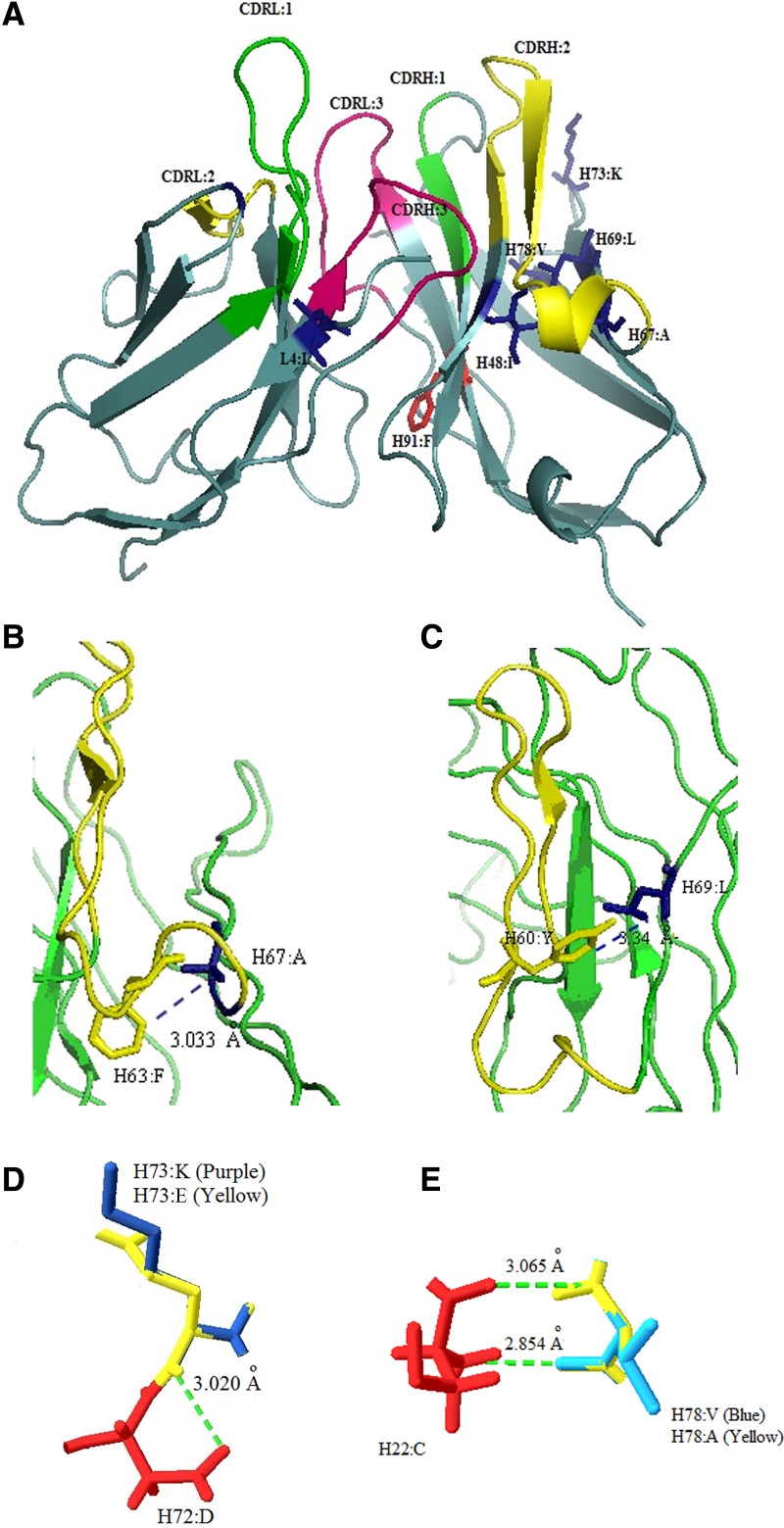
To demonstrate that some mouse residues could be replaced with human residues to make humanized scFv as close to the human sequence as possible, different Vernier zone and chain-packing residues were examined individually via molecular modeling of m5E11scFv (Fig. 3A). Molecular structure analysis revealed that AlaH:67 is positioned 3.033 Å away from PheH:63 (a CDRH2 residue) and LeuH:69 is positioned 3.340 Å away from TyrH:60 (a CDRH2 residue). Both residues thus form a weak Van der Waals interaction with these CDR residues (Fig. 3B, C). In addition, LeuL:4, an N-terminal residue of VL, formed surface contact with the antigen binding pocket(31) while IleH:48 was found to be buried in the antigen binding pocket (Fig. 3A). Both residues may have had direct contact with the antigen. The molecular model of m5E11scFv showed that PheH:91, the chain-packing residue, was buried in the antigen binding pocket (Fig. 3A). Therefore, these five mouse residues were retained in the final h5E11scFv structure. The remaining differing Vernier zone residues could be substituted with their respective human counterparts (LysH:73 to Glu; ValH:78 to Ala) due to their similar properties of being hydrophilic (LysH:73 to Glu) or hydrophobic (ValH:78 to Ala) amino acids. Interaction between the residues of interest and nearby amino acids was analyzed by superimposing mouse residues with their corresponding human counterparts. Results showed that interactions between residues were retained (Fig. 3D, E). The results implied that substitution of mouse Vernier zone residues with their respective human counterpart did not alter RMSD value (1.46 Å). Careful editing of these mouse residues allowed the engineered h5E11scFv sequence to be as close to the human sequence as possible, resulting in 95.3% amino acid sequence identity to the selected human FR sequence of VH and 98.8% amino acid sequence identity to the selected human FR sequence of VL.
FIG. 3.
Molecular structural analysis of m5E11scFv. (A) Vernier zone and chain-packing residues that are different from human consensus sequences (shown in blue and red, respectively). (B) Interaction between Vernier zone residues (AlaH:67) and amino acid residues in CDR2 (Phe;H63:F). (C) Interaction between Vernier zone residues (LeuH:69) and amino acid residues in CDR2 (Tyr;H60:Y). (D) Superimposition of human residue Glu (H73:E, yellow) onto mouse residue Lys (H73:K: purple). (E) Superimposition of human residue Ala (H78:A, yellow) onto mouse residue Val (H78:V, blue). Interactions between residues of interest and nearby amino acids are also indicated.
ScFv expression and improvement in production yield of cysteine-tagged humanized scFvs
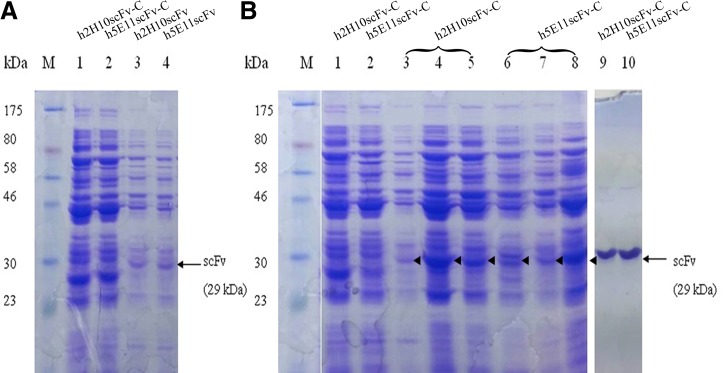
After designing the humanized scFvs, the resulting h2H10scFv and h5E11scFv sequences were subjected to gene synthesis in scFv format. The mouse and humanized scFv synthetic genes were cloned into multi-cloning sites of the pET26b+ plasmid, after the pelB signal sequence, to direct the target protein to the periplasm. Recombinant humanized scFvs were expressed in the E. coli expression system. After adding 1 mM IPTG and allowing further growth at 20°C for 18 h, soluble scFvs were successfully expressed and purified from bacterial periplasmic fraction. All expressed humanized scFvs were systemically obtained at a consistent yield of approximately 2 mg/L culture volume, with a molecular mass of 29 kDa (Fig. 4A).
FIG. 4.
(A) 12% SDS-PAGE analysis of humanized scFv expression from periplasmic extract. Lanes 1 and 2, periplasmic expression of h2H10scFv-C and h5E11scFv-C, respectively; lanes 3 and 4, periplasmic expression of h2H10scFv and h5E11scFv, respectively. (B) Effects of condition optimization on hscFv-C expression. Lanes 1 and 2, periplasmic expression of h2H10scFv-C and h5E11scFv-C, respectively; lanes 3–5, periplasmic expression of h2H10scFv-C from E. coli cultured with 0.4M sucrose, 1% Triton X-100, and 2% glycine, respectively; lanes 6–8, periplasmic expression of h5E11scFv-C from E. coli cultured with 0.4 M sucrose, 2% glycine, and 1% Triton X-100, respectively; lanes 9 and 10, purified h2H10scFv-C and h5E11scFv-C, respectively; lane M, protein molecular weight marker. Band for scFv is indicated by arrow, with expected molecular mass of 29 kDa.
For potential targeting application, both humanized scFv genes were modified by adding a cysteine residue at the C-terminus of the humanized scFv structure (hscFv-C), permitting a site-directed chemical conjugation. After induction of E. coli using the same conditions applied to non-cysteine–tagged humanized scFv, we found that yields of h2H10scFv-C and h5E11scFv-C reduced to approximately 0.2 mg/L culture (Fig. 4A). This finding was consistent with previous publications, which reported that presence of unpaired cysteine affected the yield of recombinant protein expressed in E. coli.(10,32) To improve protein production yield, conditions were optimized as follows: reduction of temperature, variation in amounts of IPTG, and addition of different concentrations of sucrose, glycine, or Triton X-100 to the culture medium after induction. Under the test conditions described in Figure 4B, the highest level of soluble hscFv-C was obtained from cultures at low temperature (15°C) with 0.125 mM IPTG and 1% Triton X-100 for 18 h. The yield of expressed protein was 4–4.5 mg/L culture. Under optimized conditions, results showed an approximate 20-fold increase in production of both hscFv-Cs, as determined by SDS-PAGE.
Bioactivity determination of engineered humanized scFv
To ensure that the targeting ability of parental MAb was retained after the humanization process, the resulting humanized scFv were evaluated for antibody binding specificity and binding affinity, as compared to parental MAb and respective mouse scFv counterpart. For h2H10scFv, binding affinity studies showed that m2H10scFv and h2H10scFv had binding affinity to EpCAM with a Kaff of 5.24 ± 0.69 × 108 M−1 and 1.90 ± 0.01 × 107 M−1, respectively. Binding affinity of h2H10scFv to EpCAM upon humanization decreased by ∼27-fold, compared with that of mouse scFv. This finding suggested that the process of humanization may have resulted in structural alteration of CDR conformation and the subsequent reduction of antigen-binding affinity. An approximate 2-fold and 60-fold reduction in Kaff, compared with parental 2H10 MAb (Kaff = 1.14 ± 0.78 × 109 M−1), were observed for m2H10scFv and h2H10scFv, respectively. Competitive ELISA was performed using varying concentrations of scFv competitors and a fixed concentration of 2H10 MAb to verify whether m2H10scFv and h2H10scFv still recognized a similar epitope on EpCAM (Fig. 5A). The results showed that binding of 2H10 MAb to EpCAM decreased in the presence of increasing amounts of scFv competitors, suggesting that both scFvs recognized the same epitope as that of their respective parental antibody. The potential for EpCAM targeting of h2H10scFv was, therefore, further demonstrated. As shown in Figure 6, HT29, an EpCAM-positive cell line, displayed high fluorescence signals after being incubated with h2H10scFv, while HEK-293T, an EpCAM-negative cell line, displayed no obvious fluorescence signal. Despite the reduction in affinity upon humanization, h2H10scFv retained epitope binding specificity and targeting ability toward EpCAM-positive cells.
FIG. 5.
Competitive ELISA for determination of antibody binding specificity of humanized scFv in comparison with parental MAb and its mouse scFv. (A) m2H10scFv (■) and h2H10scFv (●) can compete against binding of 2H10 MAb to EpCAM. (B) m5E11scFv (■) and h5E11scFv (●) can compete against binding of 5E11 MAb to HA. Experiments were performed in triplicate, with data expressed as mean ± SD.
FIG. 6.
Cell targeting ability of h2H10scFv in comparison with 2H10 MAb, as determined by confocal microscopy. HT29 and HEK-293T cells are stained with 2H10 MAb and h2H10scFv. Nuclei are labeled with Hoechst 33342 (blue).
Regarding h5E11scFv, both m5E11scFv and h5E11scFv had comparable binding affinity to hemagglutinin (HA), with a Kaff of 1.56 ± 0.15 × 107 M−1 and 1.64 ± 0.13 × 107 M−1, respectively. As shown in Figure 5B, h5E11scFv and m5E11scFv retained their epitope binding specificity, as they successfully competed with the HA binding of 5E11 MAb. In addition, h5E11scFv demonstrated targeting ability to H5N1 virus-infected cells, as reported in our recent publication.(33) However, both scFvs had lower binding affinities than their respective parental 5E11 MAb. An approximate 40-fold reduction in Kaff (5E11 MAb; Kaff- = 7.06 ± 0.6 × 108 M−1) was observed. Taken together, these results demonstrated that h5E11scFv fully retained the characteristics of m5E11scFv after humanization.
Discussion
The aim of this study was to demonstrate a concise method of generating humanized scFv from mouse hybridoma-derived antibody. Recent studies have demonstrated use of scFv as a targeting ligand for delivery of drugs or siRNAs to cancer cells and have also reported on targeting applications in virus infection models.(34,35) In this study, two promising targeting antibodies derived from mouse hybridomas were humanized—specifically, 2H10 and 5E11 MAbs. The first clone, 2H10 MAb, demonstrated selective binding against EpCAM-positive cancer cell lines. The epithelial cell adhesion molecule (EpCAM) is overexpressed in most solid cancers, while limited in normal tissues.(36) Therefore, an scFv specific to EpCAM is an attractive biological tool in cancer detection for in vivo targeting. The second clone, 5E11 MAb, recognized hemagglutinin (HA) of H5N1 influenza virus, exhibiting potent neutralizing activity against isolated H5N1 viruses. In the case of influenza virus, a HA protein is abundantly expressed on the surface of a virus-infected cell. Therefore, an scFv against HA would have practical use in delivering therapeutic agents to these virus-infected cells.(33)
Three key factors are responsible for the success of our humanization method. First, selected human templates have as much similarity as possible to the mouse FR sequences, as well as similarity in CDR canonical structures. Second, mouse residues at positions that can strongly affect the structure of CDRs are retained or back-mutated into selected human FRs to prevent affinity loss. Third, the RMSD value of the two superimposed scFv is as low as possible to facilitate CDR conformational preservation during the humanization process.
In the humanization process, it is possible to graft 6 CDRs onto human FRs, which may come from fixed framework, human immunoglobulin homology sequences, human immunoglobulin germline sequences, or human immunoglobulin consensus sequences.(31) We intentionally selected human immunoglobulin germline and human immunoglobulin consensus sequences as the human acceptor FRs to rule out possible unusual residues that may occur due to somatic mutations when fixed framework or human immunoglobulin homology sequences are used. The most identical sequence between mouse and acceptor FRs is able to retain critical residues, adversely affecting CDR conformation in the humanized scFv. Canonical structural class determines the conformation of CDRs.(37) Therefore, if the selected human sequences have similarly structured CDRs, the human acceptor FRs are likely to support the mouse CDRs. In this study, the canonical structural class of CDRs of both mouse and human sequences were identified. Specifically, four of five were identical for 2H10 and three of five were identical for 5E11. In a previous study, a humanized antibody against hepatitis B surface antigen was successfully generated with retention of high affinity, with three of five CDR canonical structures identical between mouse and selected human consensus sequences.(28)
To prevent binding affinity loss, we first focused on the preservation of all mouse residues at the Vernier zone and the chain-packing residues in both humanized scFvs. We identified the mouse residues that needed to be retained or back-mutated by comparing the primary sequence of mouse and human acceptor FRs to avoid the difficulty of identifying back mutation by computer-assisted molecular modeling. The combination of CDR grafting and preservation of the mouse Vernier zone or chain-packing residues has led to successful humanization of several antibodies.(28,38)
RMSD values of the two superimposed proteins reflect the structural differences.(39) A high RMSD value correlates with dissimilar 3-D structures, while an RMSD value of zero correlates with identical 3-D structural conformation.(40) Regarding RMSD value <1.5 Å, calculation on alpha carbon atom is proposed to be suitable for defining similarity in antibody epitopes.(41–43) We therefore attempted to design our two humanized scFvs with RMSD <1.5 Å to prevent loss of original CDR conformation and resulting loss of binding affinity. To evaluate the impact of back-mutation residues on CDR conformation, the RMSD value of mouse scFv structure, superimposed with the humanized scFv structure, was calculated. The RMSD values were found to significantly decrease after back mutations, indicating an increase in similarity between CDR conformations compared with those of mouse scFvs. We observed that, other than back mutation of Vernier zone and chain-packing residues, back mutations at positions that differ in hydrophobic or hydrophilic properties also had an influence on CDR conformational preservation, such as in h2H10scFv. However, h2H10scFv, with an RMSD of 1.6 Å, had a 27 times reduction in binding affinity after humanization. Conversely, h5E11scFv, with an RMSD of 1.4 Å, completely preserved its binding affinity compared to that of mouse scFv.
In order to improve the binding affinity of hu2H10scFv, we suggest that additional mouse residues could be back-mutated onto the designed humanized sequence to shift RMSD toward a value <1.5 Å. For example, mouse residues within approximately 5 Å of CDRs could be identified as mouse FR residues for back mutation.(44,45) It should be noted that the presence of more back-mutation sites in a humanized antibody could increase the immunogenicity of the antibodies, causing the introduction of new MHC class II epitopes.
Guided by 3-D computer modeling, some mouse Vernier zone residues could be replaced with their human counterpart to allow the humanized scFv to become closer to the human sequence, as demonstrated in h5E11scFv. For this purpose, the 3-D structure-based design methodology relied on two simple criteria. First, all mouse residues that might have direct contact with the antigen were retained to avoid disturbing any important interactions within the antigen binding pocket (AlaH:67, LeuH:69, LeuL:4, IleH:48, and PheH:91). Second, only the substitutions that would not critically affect interaction with nearby residues were selected. The substitution of a hydrophilic residue for a buried hydrophobic residue or any other substitution that would disrupt any chemical bonding was carefully avoided. Using these two criteria, two mouse Vernier zone residues were substituted with their respective human counterparts (LysH:73 to Glu and ValH:78 to Ala). The RMSD value of h5E11scFv (1.46 Å) did not change after the substitution, indicating that some mouse Vernier zone residues could be replaced with human residues without interfering with CDR conformation. As a result, the obtained h5E11scFv successfully retained the antigen-binding affinity of mouse 5E11scFv. A similar finding was reported in a previous study that focused on the development of a humanized antibody against human intercellular adhesion molecule 1 (ICAM-1), in which some mouse Vernier zone residues were replaced with human residues with no effect on antibody affinity.(20)
Notably, both the mouse and humanized scFv formats had lower binding affinity compared to their respective intact parental antibodies, with a 2-fold and 60-fold reduction in binding affinity for m2H10scFv and h2H10scFv, respectively. In the case of 5E11scFvs, binding affinity was less than that of 5E11 MAb, with an approximate 40-fold reduction in Kaff. Similar results have been consistently reported by other studies, which demonstrates that scFvs exhibit a reduction in binding affinity, as compared to that of the original whole antibody.(46–48) With many scFv molecules, this reduction in affinity develops when the format is switched from larger bivalent monoclonal antibody to smaller monovalent scFv. In most cases, scFvs have been shown to retain the antigen-binding affinity of the monovalent Fab′ fragment.(49) Formation of multimeric scFvs or performing site-directed mutagenesis within one or more CDRs were suggested as ways to increase the affinity of scFvs.(50,51)
For potential targeted delivery application, the designed humanized scFv sequences were engineered to include a cysteine amino acid at the C-terminus, providing a free thiol group to allow site-directed coupling of the scFv with other biomolecules. Our recent publication has demonstrated targeting ability of h5E11scFv-C to H5N1 virus-infected cells.(33) In that study, h5E11scFv-C has been employed as a targeting ligand by site-directed coupling to the maleimide group on the liposomes to generate immunoliposomes. The immunoliposomes were shown to specifically bind HA-expressing Sf9 cells and demonstrated enhanced siRNA transfection efficiency. The study is underway to evaluate potential targeting application of h2H10scFv-C in cancer model.
The Pel B leader sequence in the expression plasmid facilitated the transportation of the scFv into the oxidizing environment of the E. coli periplasmic space, a space where two intra-domain disulfide bonds in the scFv can take place.(52) However, previous findings have indicated that the presence of unpaired cysteine at the C-terminus affects the yield of soluble scFv production by forming covalent aggregates with other proteins containing cysteine within the periplasmic space.(10,32) To overcome this obstacle, the conditions for hscFv-C production were investigated. Under optimized conditions, we cultured E. coli at low temperature and low IPTG to sustain a reasonably low speed, resulting in the facilitation of correct scFv folding into a soluble form. Yang and colleagues(53) reported that adding glycine and Triton X-100 enhances protein secretion into the culture medium. The loss of integrity of the cellular membrane may be a potential mechanism for such an enhancement. We found that adding 1% Triton X-100 dramatically increased production of both hscFv-Cs in the periplasm, with an approximate 20-fold increase in soluble hscFv-C production being achieved. One possible explanation suggests that 1% Triton X-100 may affect the membrane integrity of the E. coli cells by increasing permeability of the inner membranes. This may result in improvement of the mechanisms associated with protein folding and export to the periplasmic space. However, low amounts of hscFv-Cs in culture supernatants were also detected in our system.
In conclusion, we successfully humanized two promising targeting antibodies. Our relatively simple humanization strategy relied on a combination of CDR grafting and preservation of mouse FR residues by primary sequence analysis. Although back mutations have to be defined individually for each antibody, we demonstrated that the combination of CDR grafting and RMSD determination made the humanization outcome both more predictable and more successful. We also demonstrated improvement in cysteine tagged scFv production, which will facilitate large-scale preparations of scFvs to meet the needs of research and clinical applications. The findings from this study may also be applied in the humanization of other antibodies derived from mouse hybridoma.
Acknowledgments
This study was supported by a research grant from the National Nanotechnology Center of the National Science and Technology Development Agency, Thailand. W.C. also received a research grant for graduate studies (Faculty of Medicine Siriraj Hospital, Mahidol University) and a Thailand Graduate Institute of Science and Technology Scholarship (National Science and Technology Development Agency, Thailand).
Author Disclosure Statement
The authors have no financial interests to disclose.
References
- 1.Nelson AL: Antibody fragments: hope and hype. MAbs 2010;2:77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Zhu X, Zhang X, Liu B, and Huang L: Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther 2010;18:1650–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dou S, Yang XZ, Xiong MH, Sun CY, Yao YD, Zhu YH, and Wang J: ScFv-decorated PEG-PLA-based nanoparticles for enhanced siRNA delivery to Her2(+) breast cancer. Adv Healthc Mater 2014;3:1792–1803 [DOI] [PubMed] [Google Scholar]
- 4.Shukra AM, Sridevi NV, Dev C, and Kapil M: Production of recombinant antibodies using bacteriophages. Eur J Microbiol Immunol (Bp);4:91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toleikis L, and Frenzel A: Cloning single-chain antibody fragments (ScFv) from hybridoma cells. Methods Mol Biol 2012;907:59–71 [DOI] [PubMed] [Google Scholar]
- 6.Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NB, and Hamid M: scFv antibody: principles and clinical application. Clin Dev Immunol 2012;1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalski M, Guindon J, Brazas L, Moore C, Entwistle J, Cizeau J, Jewett MA, and Macdonald GC: A phase II study of oportuzuMAb monatox: an immunotoxin therapy for patients with noninvasive urothelial carcinoma in situ previously treated with Bacillus Calmette-Guerin. J Urol 2012;188:1712–1718 [DOI] [PubMed] [Google Scholar]
- 8.Shernan SK, Fitch JC, Nussmeier NA, Chen JC, Rollins SA, Mojcik CF, Malloy KJ, Todaro TG, Filloon T, Boyce SW, et al. : Impact of pexelizuMAb, an anti-C5 complement antibody, on total mortality and adverse cardiovascular outcomes in cardiac surgical patients undergoing cardiopulmonary bypass. Ann Thorac Surg 2004;77:942-949; discussion, 949–950. [DOI] [PubMed] [Google Scholar]
- 9.Karwa R, and Wargo KA: EfunguMAb: a novel agent in the treatment of invasive candidiasis. Ann Pharmacother 2009;43:1818–1823 [DOI] [PubMed] [Google Scholar]
- 10.Albrecht H, Burke PA, Natarajan A, Xiong CY, Kalicinsky M, DeNardo GL, and DeNardo SJ: Production of soluble ScFvs with C-terminal-free thiol for site-specific conjugation or stable dimeric ScFvs on demand. Bioconjug Chem 2004;15:16–26 [DOI] [PubMed] [Google Scholar]
- 11.Natarajan A, Xiong CY, Albrecht H, DeNardo GL, and DeNardo SJ: Characterization of site-specific ScFv PEGylation for tumor-targeting pharmaceuticals. Bioconjug Chem 2005;16:113–121 [DOI] [PubMed] [Google Scholar]
- 12.Verhaar MJ, Keep PA, Hawkins RE, Robson L, Casey JL, Pedley B, Boden JA, Begent RH, and Chester KA: Technetium-99m radiolabeling using a phage-derived single-chain Fv with a C-terminal cysteine. J Nucl Med 1996;37:868–872 [PubMed] [Google Scholar]
- 13.Ahmadzadeh V, Farajnia S, Feizi MA, and Nejad RA: Antibody humanization methods for development of therapeutic applications. Monoclon Antib Immunodiagn Immunother 2014;33:67–73 [DOI] [PubMed] [Google Scholar]
- 14.Safdari Y, Farajnia S, Asgharzadeh M, and Khalili M: Antibody humanization methods—a review and update. Biotechnol Genet Eng Rev 2013;29:175–186 [DOI] [PubMed] [Google Scholar]
- 15.Kim JH, and Hong HJ: Humanization by CDR grafting and specificity-determining residue grafting. Methods Mol Biol 2012;907:237–245 [DOI] [PubMed] [Google Scholar]
- 16.Saldanha JW, Martin AC, and Leger OJ: A single backmutation in the human kIV framework of a previously unsuccessfully humanized antibody restores the binding activity and increases the secretion in cos cells. Mol Immunol 1999;36:709–719 [DOI] [PubMed] [Google Scholar]
- 17.Ewert S, Honegger A, and Pluckthun A: Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods 2004;34:184–199 [DOI] [PubMed] [Google Scholar]
- 18.Foote J, and Winter G: Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol 1992;224:487–499 [DOI] [PubMed] [Google Scholar]
- 19.Hou S, Li B, Wang L, Qian W, Zhang D, Hong X, Wang H, and Guo Y: Humanization of an anti-CD34 monoclonal antibody by complementarity-determining region grafting based on computer-assisted molecular modelling. J Biochem 2008;144:115–120 [DOI] [PubMed] [Google Scholar]
- 20.Luo GX, Kohlstaedt LA, Charles CH, Gorfain E, Morantte I, Williams JH, and Fang F: Humanization of an anti-ICAM-1 antibody with over 50-fold affinity and functional improvement. J Immunol Methods 2003;275:31–40 [DOI] [PubMed] [Google Scholar]
- 21.Villani ME, Morea V, Consalvi V, Chiaraluce R, Desiderio A, Benvenuto E, and Donini M: Humanization of a highly stable single-chain antibody by structure-based antigen-binding site grafting. Mol Immunol 2008;45:2474–2485 [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Raifu M, Howard M, Smith L, Hansen D, Goldsby R, and Ratner D: Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J Immunol Methods 2000;233:167–177 [DOI] [PubMed] [Google Scholar]
- 23.Eswarakumar VP, Raja MC, and Muthukkaruppan VR: RT-PCR cloning and characterization of mouse immunoglobulin variable domains with high affinity for HLA-DR antigens. Immunogenetics 1997;46:249–250 [DOI] [PubMed] [Google Scholar]
- 24.Kabat EA, Wu TT, Perry HM, Gottesman KS, and Foeller: Sequences of Proteins of Immunological Interest, 5th ed. U.S. Dept. of Health and Human Services, Bethesda, MD, 1991 [Google Scholar]
- 25.Beatty JD, Beatty BG, and Vlahos WG: Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J Immunol Methods 1987;100:173–179 [DOI] [PubMed] [Google Scholar]
- 26.Makabe K, Nakanishi T, Tsumoto K, Tanaka Y, Kondo H, Umetsu M, Sone Y, Asano R, and Kumagai I: Thermodynamic consequences of mutations in vernier zone residues of a humanized anti-human epidermal growth factor receptor murine antibody, 528. J Biol Chem 2008;283:1156–1166 [DOI] [PubMed] [Google Scholar]
- 27.Chothia C, Novotny J, Bruccoleri R, and Karplus M: Domain association in immunoglobulin molecules. The packing of variable domains. J Mol Biol 1985;186:651–663 [DOI] [PubMed] [Google Scholar]
- 28.Tiwari A, Khanna N, Acharya SK, and Sinha S: Humanization of high affinity anti-HBs antibody by using human consensus sequence and modification of selected minimal positional template and packing residues. Vaccine 2009;27:2356–2366 [DOI] [PubMed] [Google Scholar]
- 29.Monera OD, Sereda TJ, Zhou NE, Kay CM, and Hodges RS: Relationship of sidechain hydrophobicity and alpha-helical propensity on the stability of the single-stranded amphipathic alpha-helix. J Pept Sci 1995;1:319–329 [DOI] [PubMed] [Google Scholar]
- 30.Kyte J, and Doolittle RF: A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982;157:105–132 [DOI] [PubMed] [Google Scholar]
- 31.Lo BK: Antibody humanization by CDR grafting. Methods Mol Biol 2004;248:135–159 [DOI] [PubMed] [Google Scholar]
- 32.Schmiedl A, Breitling F, Winter CH, Queitsch I, and Dubel S: Effects of unpaired cysteines on yield, solubility and activity of different recombinant antibody constructs expressed in E. coli. J Immunol Methods 2000;242:101–114 [DOI] [PubMed] [Google Scholar]
- 33.Khantasup K, Kopermsub P, Chaichoun K, and Dharakul T: Targeted small interfering RNA-immunoliposomes as a promising therapeutic agent against highly pathogenic avian influenza A (H5N1) virus infection. Antimicrob Agents Chemother 2014;58:2816–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirollo KF, Zon G, Rait A, Zhou Q, Yu W, Hogrefe R, and Chang EH: Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum Gene Ther 2006;17:117–124 [DOI] [PubMed] [Google Scholar]
- 35.Wen WH, Liu JY, Qin WJ, Zhao J, Wang T, Jia LT, Meng YL, Gao H, Xue CF, Jin BQ, et al. : Targeted inhibition of HBV gene expression by single-chain antibody mediated small interfering RNA delivery. Hepatology 2007;46:84–94 [DOI] [PubMed] [Google Scholar]
- 36.Munz M, Baeuerle PA, and Gires O: The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res 2009;69:5627–5629 [DOI] [PubMed] [Google Scholar]
- 37.Al-Lazikani B, Lesk AM, and Chothia C: Standard conformations for the canonical structures of immunoglobulins. J Mol Biol 1997;273:927–948 [DOI] [PubMed] [Google Scholar]
- 38.Shembekar N, Mallajosyula VV, Chaudhary P, Upadhyay V, Varadarajan R, and Gupta SK: Humanized antibody neutralizing 2009 pandemic H1N1 virus. Biotechnol J 2014;9:1594–1603 [DOI] [PubMed] [Google Scholar]
- 39.Maiorov VN, and Crippen GM: Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J Mol Biol 1994;235:625–634 [DOI] [PubMed] [Google Scholar]
- 40.Carugo O, and Pongor S: A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci 2001;10:1470–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker D, and Sali A: Protein structure prediction and structural genomics. Science 2001;294:93–96 [DOI] [PubMed] [Google Scholar]
- 42.Mader A, and Kunert R: Humanization strategies for an anti-idiotypic antibody mimicking HIV-1 gp41. Protein Eng Des Sel 2010;23:947–954 [DOI] [PubMed] [Google Scholar]
- 43.Zhang D, Chen CF, Zhao BB, Gong LL, Jin WJ, Liu JJ, Wang JF, Wang TT, Yuan XH, and He YW: A novel antibody humanization method based on epitopes scanning and molecular dynamics simulation. PLoS One 2013;8:e80636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, Wang H, Zhang D, Qian W, Hou S, Shi S, Zhao L, Kou G, Cao Z, Dai J, et al. : Construction and characterization of a high-affinity humanized SM5-1 monoclonal antibody. Biochem Biophys Res Commun 2007;357:951–956 [DOI] [PubMed] [Google Scholar]
- 45.Chiu WC, Lai YP, and Chou MY: Humanization and characterization of an anti-human TNF-alpha murine monoclonal antibody. PLoS One 2011;6:e16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo JQ, You SY, Li L, Zhang YZ, Huang JN, and Zhang CY: Construction and high-level expression of a single-chain Fv antibody fragment specific for acidic isoferritin in Escherichia coli. J Biotechnol 2003;102:177–189 [DOI] [PubMed] [Google Scholar]
- 47.Muramatsu H, Yoshikawa K, Hayashi T, Takasu S, Kawada Y, Uchida K, Sato S, Takahashi T, Saga S, and Ueda R: Production and characterization of an active single-chain variable fragment antibody recognizing CD25. Cancer Lett 2005;225:225–236 [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi D, Watanabe N, Yamauchi N, Okamoto T, Tsuji N, Sasaki H, Sato T, and Niitsu Y: Heat-induced apoptosis via caspase-3 activation in tumor cells carrying mutant p53. Int J Hyperthermia 2000;16:471–480 [DOI] [PubMed] [Google Scholar]
- 49.Batra SK, Jain M, Wittel UA, Chauhan SC, and Colcher D: Pharmacokinetics and biodistribution of genetically engineered antibodies. Curr Opin Biotechnol 2002;13:603–608 [DOI] [PubMed] [Google Scholar]
- 50.Kuan CT, Wikstrand CJ, Archer G, Beers R, Pastan I, Zalutsky MR, and Bigner DD: Increased binding affinity enhances targeting of glioma xenografts by EGFRvIII-specific scFv. Int J Cancer 2000;88:962–969 [DOI] [PubMed] [Google Scholar]
- 51.Kortt AA, Dolezal O, Power BE, and Hudson PJ: Dimeric and trimeric antibodies: high avidity scFvs for cancer targeting. Biomol Eng 2001;18:95–108 [DOI] [PubMed] [Google Scholar]
- 52.Oelschlaeger P, Lange S, Schmitt J, Siemann M, Reuss M, and Schmid RD: Identification of factors impeding the production of a single-chain antibody fragment in Escherichia coli by comparing in vivo and in vitro expression. Appl Microbiol Biotechnol 2003;61:123–132 [DOI] [PubMed] [Google Scholar]
- 53.Yang J, Moyana T, MacKenzie S, Xia Q, and Xiang J: One hundred seventy-fold increase in excretion of an FV fragment-tumor necrosis factor alpha fusion protein (sFV/TNF-alpha) from Escherichia coli caused by the synergistic effects of glycine and triton X-100. Appl Environ Microbiol 1998;64:2869–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]