Abstract
Toll-like receptor 4 (TLR4), the receptor for the bacterial product endotoxin, is subject to multiple points of regulation at the levels of signaling, biogenesis, and trafficking. Dysregulation of TLR4 signaling can cause serious inflammatory diseases, such as sepsis. We found that the p24 family protein TMED7 (transmembrane emp24 protein transport domain containing 7) is required for the trafficking of TLR4 from the endoplasmic reticulum to the cell surface through the Golgi. TMED7 formed a stable complex with the ectodomain of TLR4, an interaction that required the coiled-coil and GOLD domains, but not the cytosolic, COP II sorting motif, of TMED7. Depletion of TMED7 reduced TLR4 signaling mediated by the adaptor protein MyD88, but not that mediated by the adaptor proteins TRAM and TRIF. Truncated forms of TMED7 lacking the COP II sorting motif or the transmembrane domain were mislocalized and resulted in constitutive activation of TLR4 signaling. Together, these results support the hypothesis that p24 proteins perform a quality control step by recognizing correctly folded anterograde cargo, such as TLR4, in early secretory compartments and facilitating the translocation of this cargo to the cell surface.
INTRODUCTION
Toll-like receptor 4 (TLR4) is a key regulator of innate immunity and inflammation. TLR4 is activated by the complex and heterogeneous glycolipid lipolysaccharide (LPS), which is present in the outer membrane of Gram negative bacteria (1). Although TLR4 is a key element of host defense against Gram negative pathogens, dysregulation of TLR4 signaling causes endotoxic shock, a severe condition that leads to multi-organ failure and death. Because of its importance in innate immunity and disease, the activity of TLR4 is highly regulated, not only by positive and negative effectors of its signaling pathways, but also at the levels of biosynthesis and trafficking.
TLR4 functions at the cell surface and also signals from early endosomes (2, 3). The trafficking events that accompany the activation of TLR4 are fairly well understood. Stimulation of TLR4 by LPS results in internalization of the receptor into early endosomes, a process that requires the accessory protein CD14, the guanosine triphosphatase (GTPase) Rab, and potentially signaling by the tyrosine kinase Syk (4-6). TLR4 is unusual because it promotes two signaling pathways. The first, which is mediated by the adaptor protein myeloid differentiation marker 88 (MyD88), leads to activation of the transcription factor nuclear factor κB (NF-κB) and proinflammatory responses, whereas the second, which is mediated by the adaptors TRAM (TRIF related adaptor molecule) and TRIF (TIR domain containing adaptor protein inducing interferon β), leads to the activation of interferon response factor 3 (IRF3) and IRF7 (7). Evidence suggests that the latter pathway is only active after internalization of TLR4. In contrast, the trafficking processes that accompany the biosynthesis of TLR4 are less well-characterized. Two chaperone molecules, gp96 and PRAT4A (protein associated with TLR4) , are required for proper processing of TLR4 in the endoplasmic reticulum (ER), which leads to the secretion of the receptor to the plasma membrane (8-10). The association of TLR4 with MD2 (myeloid differentiation factor 2) in the ER is also critical for the correct glycosylation of TLR4, its secretion to the plasma membrane, and hence its responsiveness to LPS (11-14). The involvement of other components of the secretory pathway, especially those related to vesicular trafficking, however, is relatively unclear. A study showed that Rab10 is responsible for transporting TLR4 from the Golgi to the plasma membrane in response to the stimulation of bone marrow-derioved macrophages and RAW264.7 cells with LPS (15).
The trafficking of cell-surface transmembrane proteins to the plasma membrane is initiated by translocation and folding of the protein in the ER. Proteins destined for secretion rather than for residence in the ER are then selectively packaged into vesicles for transport to the cis-Golgi. This process is dependent on the ability of the cargo protein to recruit COP II (coat protein complex II), which leads to their assembly around the budding membrane before vesicle formation (16). This recruitment is mediated by the presence of the COP II–binding motif of di-phenylalanine in the cytosolic tail of the cargo protein (17-19). However, soluble cargo proteins (and indeed many transmembrane proteins, including TLR4) do not have this cytosolic diphenylalanine motif. Instead, these cargo molecules are selected by transmembrane adaptor proteins that have recognition domains that are exposed in the lumen of the ER, as well as short cytoplasmic tails that include the diphenylalanine sorting motif for COP II and a dibasic signal for COP I–dependent retrograde transport back to the ER from the Golgi (20).
One important family of transmembrane adaptor proteins includes the p24 proteins, alternatively called TMED (transmembrane emp24 protein transport domain containing) proteins in vertebrates. There are ten family members in mammals, and they are conserved across species. TMEDs consist of golgi dynamics (GOLD) and coiled-coil domains in the lumen of the ER, a single transmembrane spanning region, and a short cytosolic tail with COP II–binding or COP I–binding motifs, or both (21, 22). TMEDs form homodimers and heterodimers and are found in the early secretory pathway of the ER, the ER Golgi intermediate compartment (ERGIC), the cis-Golgi, as well as COP I– and COP II–coated vesicles (19, 23-26). In yeast, loss of several p24 proteins leads to defects in the secretion of several proteins and the formation of secretory vesicles, as well as activation of the unfolded protein response (UPR), but these effects are not lethal (27-29). In mice, deletion of certain TMEDs leads to defective embryogenesis, which can be lethal (30, 31). Although it is clear that TMEDs have a fundamental cell biological function in the selection of secretory cargo, little is known about the nature of the cargo molecules or the molecular mechanisms by which these molecules are recognized in the ER and dissociated in the Golgi (22). Here, we showed that the pattern-recognition receptor TLR4 is a cargo molecule for TMED7, which tightly regulates trafficking of the receptor to the cell surface. Dysregulation of TMED7-mediated transport led to the constitutive activation of the TLR4 signaling pathway.
RESULTS
TMED7 is a type I membrane protein with a luminal N-terminal GOLD domain
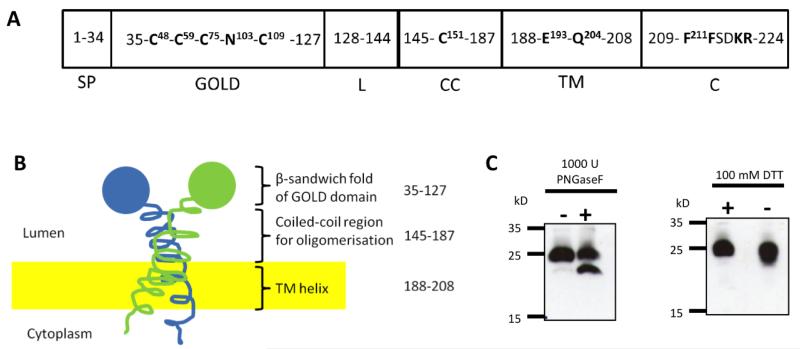
Previous reports suggested that TMED7 is localized on the cytosolic surface of the plasma membrane (32); however, those studies contradict bioinformatic analysis that predicts that TMED7 is a secreted membrane protein with a type I topology and a short cytosolic domain at the C-terminus (Fig. 1, A and B and fig. S1, A and B). To resolve this point, we investigated whether TMED7 contains N-linked glycans and disulphide bonds, posttranslational modifications that are added to proteins in the lumen of the ER during biosynthesis. Treatment of transfected human embryonic kidney (HEK) 293T cell extracts with the glycosidase PNGaseF led to the formation of a species of FLAG-tagged TMED7 (TMED7-FLAG) that migrated more quickly by SDS-PAGE than did TMED7-FLAG from untreated cells (Fig. 1C). This result suggests that TMED7 is glycosylated, most likely at Asn103 in the GOLD domain, the only predicted N-linked glycosylation site found in the protein with the consensus sequence of Asn-Xaa-Thr, which is conserved in TMED7 across various species (fig. S1C). Treatment of cell extracts with PNGaseF also led to a change in the electrophoretic mobility of TLR4, which is heavily glycosylated, but not of TRAM, which is not glycosylated (fig. S2A). This finding confirms the observation made by Fullekrug et al. who showed that native TMED7 (gp27) contains complex glycans and terminal sialic acid residues (24).
Fig. 1. Characterization of the organization of the TMED7 protein.
(A) Arrangement of the domains of TMED7 based on various prediction tools and previous studies (19, 24, 46, 52) with amino acid residue position numbers indicated in the domain boxes. Cys48 and Cys109 are conserved in all members of the p24 family across all species studied, whereas the other Cys residues are not (22). Asn103 is predicted to be a site of N-glycosylation. Glu193 and Gln204 are sorting signals within the transmembrane domain that modulate the vesicular trafficking activity of TMED7 (33). The two phenylalanine residues in the cytoplasmic tail form a motif that is involved in binding to COP II (19). SP, signal peptide; GOLD, Golgi dynamics domain; L, linker; CC, coiled-coil domain; TM, transmembrane domain; C, cytoplasmic tail. (B) Representation of the domain organization and topology of TMED7, with its N-terminal GOLD domain located within the lumen of the ER. Amino acid residues are indicated on the right. (C) HEK 293T cells were transiently transfected with plasmid encoding HA-tagged TMED7 (TMED7-HA). 48 hours later, whole-cell lysates were collected and were left untreated or were treated with PNGaseF, to detect the glycosylation state of TMED7, or with DTT, to detect the presence of any disulfide bridges in the protein. Lysates were analyzed by Western blotting with an anti-HA antibody. Blots are representative of two independent experiments.
Multiple sequence alignments of p24 proteins revealed that there are two conserved cysteine residues within the GOLD domain that may be involved in the formation of a disulfide bridge (21). To test this hypothesis, we transfected HEK 293T cells with plasmid encoding hemagglutinin (HA)-tagged TMED7 (TMED7-HA) and analyzed the protein under reducing and nonreducing conditions. The band corresponding to TMED7 migrated faster under nonreducing conditions than it did under reducing conditions, which is a characteristic of the oxidized denatured state (Fig. 1C). Finally, we transfected Hi5 insect cells with plasmids encoding recombinant human TMED7 proteins containing either its native signal peptide (SP) or a fusion signal peptide (SP-CC and PP-CC, respectively), and analyzed them by Edman degradation, which revealed that a hydrophobic N-terminal signal sequence was cleaved before the residue Ser35 (fig. S2, B and C) (19). Together, these data support the predicted membrane topology of TMED7 (Fig. 1B).
TMED7 oligomers physically interact with TLR4 in the Golgi
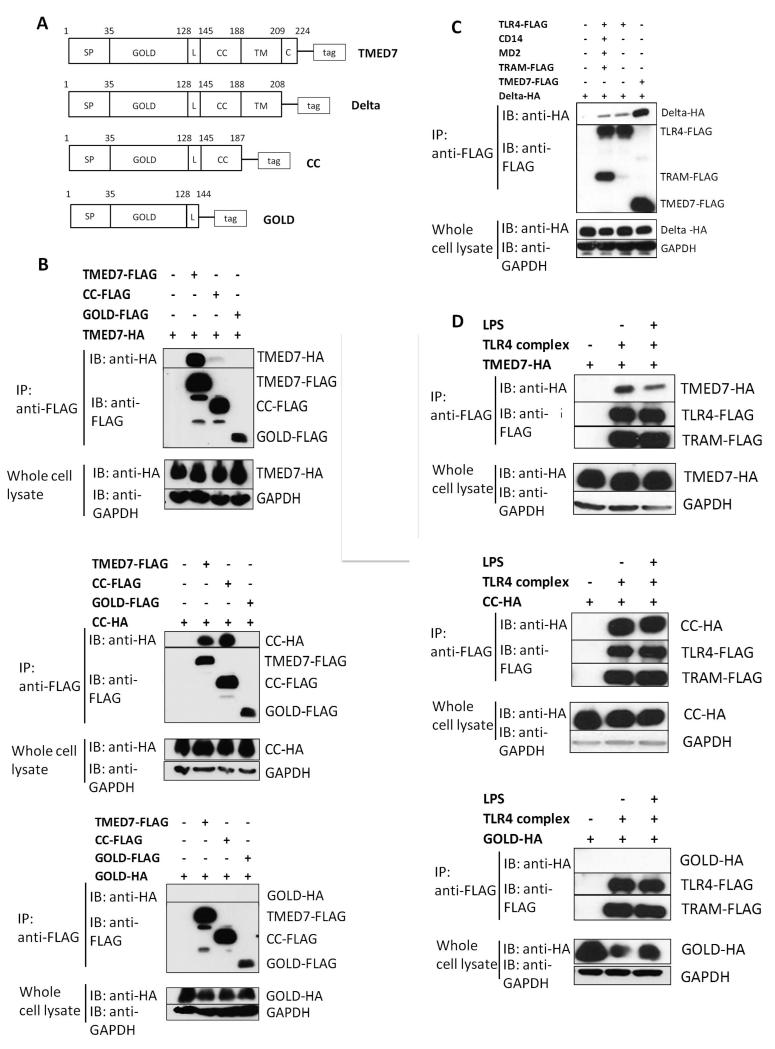
We next investigated whether TMED7 formed dimers or oligomers of itself and could physically interact with TLR4. We transfected HEK 293T cells with plasmids encoding full-length TMED7 or three truncated mutants of TMED7 that contained C-terminal HA or FLAG tags (Fig. 2A). The Delta truncated mutant of TMED7 is deficient in the cytoplasmic tail region; the CC truncated mutant is missing both the transmembrane and cytosolic tail regions; whereas the GOLD truncated mutant of TMED7 is deficient in the cytosolic, transmembrane, and coiled coil regions (Fig. 2A). Samples from transfected cells were subjected to immunoprecipitation with an anti-FLAG antibody and then were analyzed by Western blotting with an anti-HA antibody to detect any HA-tagged proteins that formed complexes. We found that full-length TMED7 and the Delta and CC constructs, but not the GOLD construct, formed oligomers between themselves and each other (Fig. 2, B and C). This suggests that the coiled-coil domain, which is lacking in the GOLD construct, is required for oligomerization. Analysis of lysates from THP-1 cells (a macrophage cell line) transduced with lentiviruses expressing the tagged TMED7 constructs showed that TMED7 formed oligomers with itself, a process that required the coiled-coil domain (fig. S3A).
Fig. 2. Oligomerization of TMED7 is crucial for its interaction with TLR4.
(A) Outline of the constructs of full-length TMED7 and of its truncated mutants: Delta, CC, and GOLD. Each construct includes a C-terminal tag: FLAG or HA. (B) Analysis of the ability of TMED7 and its truncated mutants to form oligomers. HEK 293T cells were transiently transfected with plasmids encoding the indicated constructs, and whole-cell lysates were incubated with anti-FLAG M2 magnetic beads to immunoprecipitate (IP) FLAG-tagged recombinant proteins. Samples were analyzed by Western blotting (IB) with anti-HA antibody to detect the interacting HA-tagged proteins and with an anti-FLAG antibody to detect the presence of the immunoprecipitated proteins. GAPDH in whole-cell lysates was used as a loading control to ensure that IPs were performed with equivalent amounts of starting material. (C) Analysis of the ability of TMED7 and its truncated mutants to interact with the TLR4 complex. HEK 293T cells were transiently transfected with plasmids encoding components of the TLR4 complex and the indicated TMED7 constructs. 48 hours later, cells were left untreated or were treated with LPS (100 ng/ml) for 30 min as indicated. Whole-cell lysates were then subjected to immunoprecipitation and Western blotting analysis as described for (B). (D) HEK 293T cells were transiently transfected with plasmids encoding the indicated constructs. 48 hours later, whole-cell lysates were subjected to immunoprecipitation and Western blotting analysis as described for (B). Western blots in all panels are representative of three independent experiments.
To test whether the TLR4 complex physically interacted with TMED7 or its truncation mutants, we transfected HEK 293T cells with plasmids encoding FLAG-TLR4, CD14, MD2, or TRAM-FLAG (TLR4 complex) together with plasmids encoding HA-tagged full-length TMED7 or its truncated mutants. FLAG-tagged proteins were immunoprecipitated and samples were analyzed by Western blotting with an anti-HA antibody to detect associated proteins. We found that full-length TMED7 and the Delta and CC truncation mutants co-immunoprecipitated with the TLR4 complex; however, the GOLD truncation mutant, which lacks the coiled-coil motif did not (Fig. 2, C and D). TLR3 which localizes in endosomal compartments did not interact with TMED7 suggesting that TEMED7 selects TLR4 specifically (fig. S3B). Treatment of the transfected HEK 293T cells with LPS (100 ng/ml) reduced the amount of full-length TMED7 that coimmunoprecipitated with TLR4, but had no effect on the amount of the CC construct that was pulled down (Fig. 2D), which suggests that LPS might stimulate the dissociation of TLR4 from TMED7.
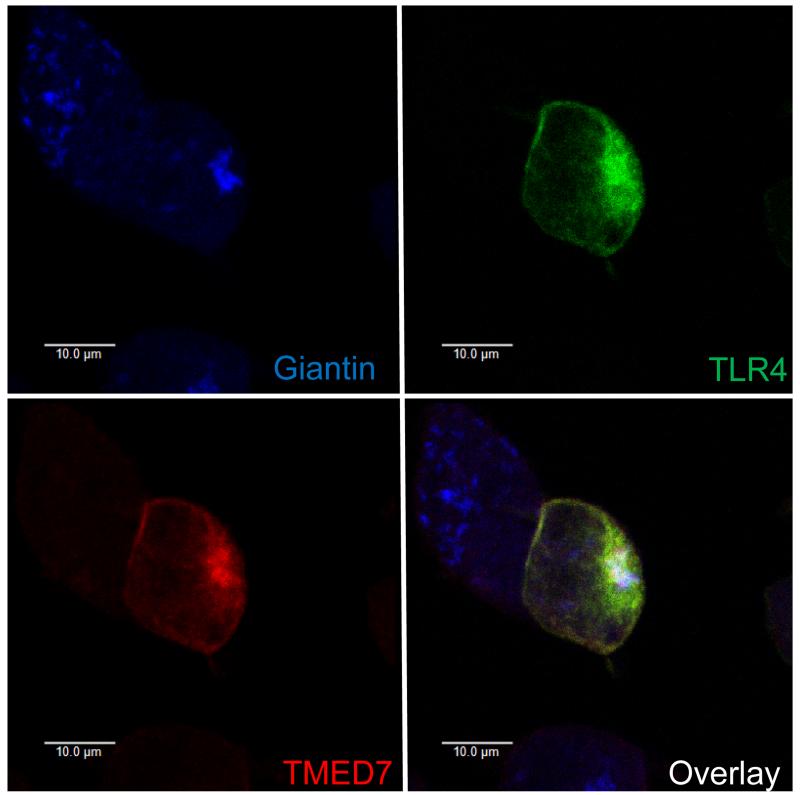
Confocal microscopic analysis of unstimulated transfected HEK 293T cells revealed that the interaction between TLR4 and TMED7 occurred in an intracellular compartment containing giantin, which is a marker for the Golgi (Fig. 3). In transduced THP-1 cells, TLR3 did not co-localize with TMED7 in the Golgi, which suggests that TMED7 may recognize TLR4 specifically (fig. S4A). Together, these data suggest that the coiled-coil domain of TMED7 mediates the formation of homodimers and heterodimers, and that TLR4 binds to dimeric or oligomeric TMED7 in the Golgi in unstimulated cells. It is possible that the interaction between TMED7 and TLR4 also requires the hetero-dimerization of TMED7 with TMED2, TMED9, or TMED10, which should be explored further (24).
Fig. 3. TLR4 and TMED7 colocalize in HEK 293T cells in the absence of LPS.
HEK 293T cells were transiently transfected with plasmids encoding TMED7-HA, TLR4-Citrine, MD2, CD14, and TRAM. Forty-eight hours after transfection, cells were permeabilized and TMED7-HA was labeled with Alexa Fluor 633 (red), and giantin was detected with Alexa Fluor 405 (blue). Cells were then analyzed by confocal microscopy. Images are representative of two independent experiments, in each of which ~20 cells were observed to have similar patterns of TMED7 and TLR4 colocalization. Scale bar: 10 μm.
Constitutive and ligand induced TLR4 signaling requires TMED7
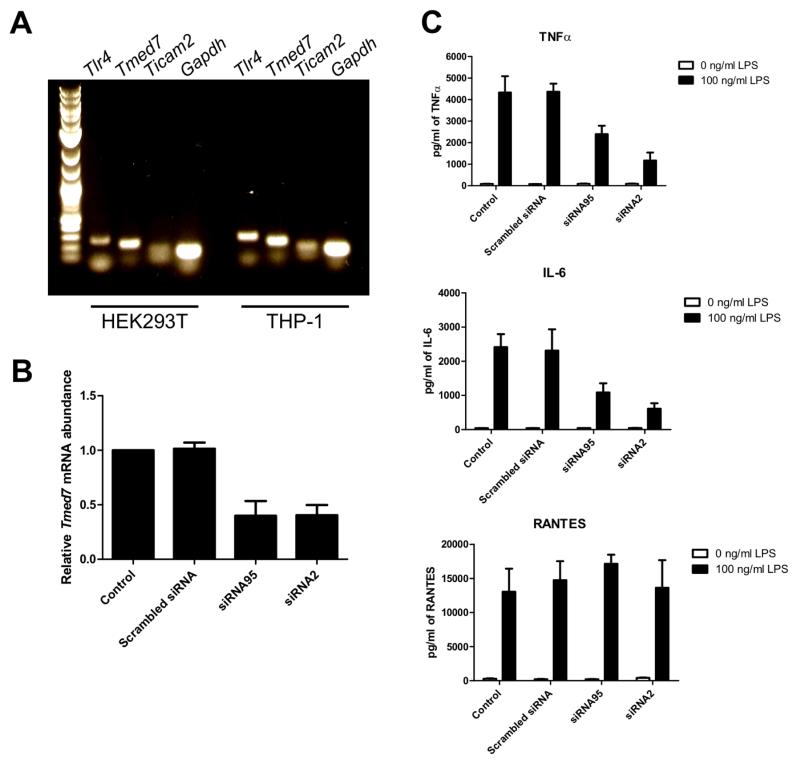
Our earlier results suggest that TMED7 and TLR4 form a stable complex in the Golgi of unstimulated cells. Previous studies showed that TMED7 is found in various early secretory compartments, including the ER, ER-Golgi intermediate compartments, and the Golgi (24, 26). Together, these studies suggest that TMED7 may recognize TLR4 in the ER as cargo for transport to the Golgi. We therefore asked whether depletion of TMED7 affected LPS-induced signaling by TLR4. We detected TMED7 mRNA in HEK 293 cells and THP-1 cells (Fig. 4A). Transfection of HEK 293 cells or THP-1 cells with TMED7-specific siRNAs resulted in a substantial reduction in the abundance of TMED7 mRNA, but a scrambled control siRNA had no effect (Fig. 4B and fig. S5A). Depletion of TMED7 in THP-1 cells with siRNA resulted in the cells secreting reduced amounts of the cytokines interleukin-6 (IL-6) and tumor necrosis factor α (TNF-α), but not RANTES, in response to LPS compared to cells transfected with nonspecific siRNA (Fig. 4C). Luciferase reporter assays performed in transfected HEK 293 cells showed that siRNA-dependent knockdown of Tmed7 resulted in reduced activation of NF-κB (MyD88-dependent), but not IFN-β (TRIF/TRAM-dependent), in response to LPS, providing further evidence that only the MyD88-dependent pathway was affected by TMED7 knockdown (fig. S5, B and C). In contrast, depletion of TMED7 did not affect TLR3 or TLR5 signaling induced by poly I:C and flagellin, respectively (fig. S5, D and E).
Fig. 4. Knockdown of TMED7 in THP-1 cells results in reduced LPS-dependent production of IL-6 and TNF-α.
(A) Total RNA isolated from HEK 293T cells and THP-1 cells was purified, and complementary DNA (cDNA) was synthesized by reverse transcription. The cDNA samples were used as templates for the detection of cDNAs for the indicated genes by PCR with specific primers. The gel is representative of three independent experiments. (B) THP-1 cells were left untreated (Control) or were transfected with the indicated siRNA duplexes (5 nM). Three days after transfection, cells were analyzed by quantitative PCR (qPCR) to determine the abundance of Tmed7 mRNA normalized to that of Actb mRNA and relative to the normalized abundance of Tmed7 mRNA in control cells, which was set at 1. Data are means ± SEM from three independent experiments. (C) Upon differentiation of THP-1 cells to macrophage-like cells, the cells were left untransfected (Control) or were transfected with the indicated siRNA duplexes (5 nM). Three days later, cells were left untreated or were stimulated with LPS (100 ng/ml) for 24 hours. Cell culture medium was then analyzed by ELISA to measure the amounts of TNF-α (top), IL-6 (middle), and RANTES (bottom) secreted by the cells. Data are means ± SD of three replicate samples from a single experiment and are representative of three independent experiments.
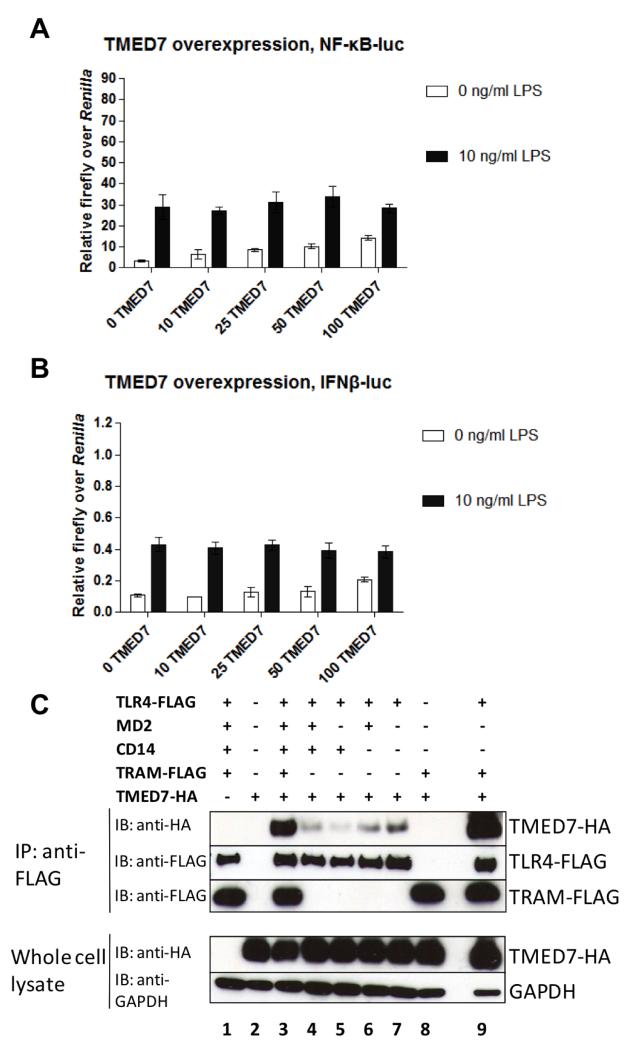
We also performed reciprocal assays by increasing the abundance of TMED7 and examining its effect on TLR4 signaling. As the amount of TMED7-encoding plasmid used to transfect HEK 293 cells was increased, the extent of basal signaling in the absence of LPS increased for both the MyD88- and TRIF/TRAM-dependent pathways (Fig. 5, A and B). In contrast, LPS-dependent signaling was unaffected by the increased TMED7 abundance (Fig. 5, A and B), which suggests that TMED7 acts upstream of the activation of TLR4 by LPS. The responses seen in this assay were specific for LPS-dependent activation of TLR4. Transfection of HEK 293 cells with plasmid encoding the GOLD TMED7 construct did not cause a similar increase in the constitutive activity of TLR4 (fig S5). Thus, the increased basal activity of TLR4 was not caused by transfecting the cells with plasmid DNA (fig. S5, A and B). Moreover, the NF-κB– and IFN-β–dependent luciferase activities required the presence of all TLR4 signaling components (fig. S5, C to E). Finally, full-length TMED7 and the Delta and CC truncation mutants did not alter the activity of TLR3 or TLR5 upon their stimulation in transiently transfected HEK 293 cells (fig. S5, G and H). Together, these data suggest that TMED7 promotes the ligand-independent activation of TLR4 signaling. To explore this finding further, we determined which components of the TLR4 complex were required to interact with TMED7. Co-immunoprecipitation experiments showed that TLR4, alone or in combination with the co-receptors CD14 and MD-2, physically interacted with TMED7; however, the extent of binding was variable, with less TMED7 associated in the presence of CD14 than in its absence (Fig. 5C). The reason for these differences is unclear. The TLR4 adaptor protein TRAM did not associate with TMED7, but it substantially enhanced the binding of TMED7 to the TLR4 complex and to TLR4 alone (Fig. 5C).
Fig. 5. TMED7 causes TLR4 signaling in the absence of LPS.
(A) TLR4 alone is sufficient to physically interact with TMED7. HEK 293T cells were transiently transfected with plasmids encoding the indicated constructs. Whole-cell lysates were prepared from unstimulated cells and were incubated with anti-FLAG M2 magnetic beads to immunoprecipitate FLAG-tagged recombinant proteins. Samples were then analyzed by Western blotting with an anti-HA antibody, to detect the interacting HA-tagged proteins, as well as an anti-FLAG antibody, to detect the immunoprecipitated proteins. GAPDH was used as a loading control. Western blots are representative of three independent experiments. (B and C) HEK 293T cells were transfected with reporter plasmids for (B) NF-κB or (C) IFN-β together with the indicated amounts (ng) of plasmid encoding TMED7-HA. 48 hours after transfection, cells were left untreated or were treated with LPS (10 ng/ml) for 6 hours and then were subjected to luciferase assays as described in the Materials and Methods. Data are the mean firefly luciferase activity normalized to Renilla luciferase activity from a single experiment, which is representative of five independent experiments. Error bars represent the SD of three technical replicates.
Regulatory roles of the cytoplasmic tail and transmembrane domain of TMED7 in TLR4 signaling
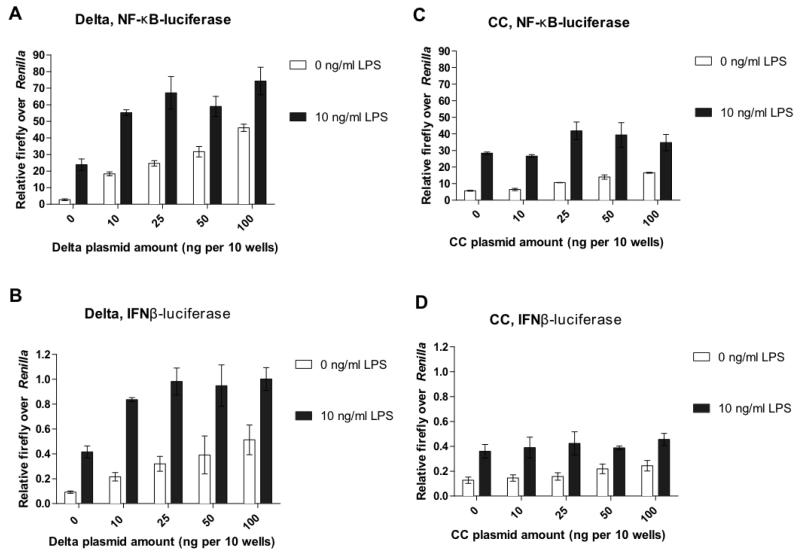
Previous studies showed that the COP I– and COP II–binding motifs in the cytoplasmic tail of TMED7, as well as the conserved Glu193 and Gln204 residues in the transmembrane domain of p24 proteins, determine the direction of trafficking and the rate of recycling of the proteins within the early secretory pathway (17, 19, 33). We therefore investigated whether these regions could also affect TLR4 signaling. We performed reporter assays with the truncated mutants of TMED7 described earlier (Fig. 2A). Transfection of HEK 293T cells with plasmid encoding the Delta TMED7 truncation mutant increased constitutive signaling through the MyD88 and TRIF/TRAM pathways by ~20-fold, but a residual amount of LPS-inducible signaling was also detected (Fig. 6, A and B). The effect of the Delta mutant was greater than that of full-length TMED7 (Fig. 5, A and B). Expression of the CC truncation mutant also enhanced the constitutive activities of both pathways to a similar extent to that of full-length TMED7 (Fig. 6, C and D), whereas expression of the TMED7 GOLD mutant did not have any effect, which was expected because this truncation mutant does not interact with TLR4 (fig. S6, A and B).
Fig. 6. Roles of the transmembrane domain and cytosolic tail of TMED7 in TLR4 signaling.
(A to D) HEK 293T cells were transfected with reporter plasmids for (A and C) NF-κB or (B and D) IFN-β together with the indicated amounts (ng) of plasmid encoding (A and B) the HA-tagged Delta mutant or (C and D) the HA-tagged CC mutant. 48 hours after transfection, cells were left untreated or were treated with LPS (10 ng/ml) for 6 hours and then were subjected to luciferase assays as described in the Materials and Methods. Data are the mean firefly luciferase activity normalized to Renilla luciferase activity from a single experiment, which is representative of five independent experiments. Error bars represent the SD of three technical replicates.
Roles of the cytoplasmic tail and transmembrane domain of TMED7 in its intracellular localization
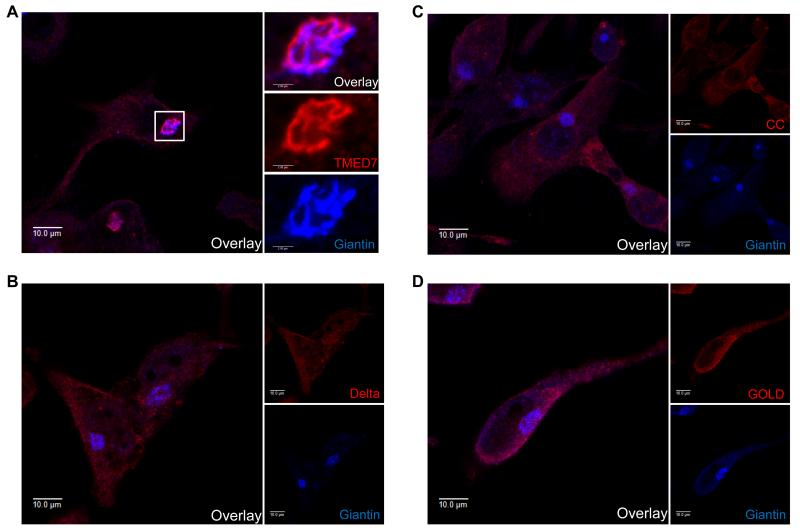
Next, we wanted to address the link between the role of TMED7 in the intracellular trafficking of TLR4 and its signaling activity. First, we confirmed the intracellular localization of TMED7 shown by previous studies (19, 24-26), and we determined whether the COP II sorting motif (Phe211Phe212) in the cytoplasmic region of TMED7 was required for this localization. We transduced THP-1 cells with lentiviruses expressing full-length and truncated TMED7 constructs and visualized the localization of these proteins and the Golgi marker giantin by immunofluorescent microscopy. As previously reported, full-length TMED7 was predominantly present in the Golgi, because it co-localized with giantin (Fig. 7A). In contrast, the Delta, CC, and GOLD truncation mutants were not concentrated in the Golgi, but adopted a reticular-like expression throughout the cell (Fig. 7, B to D). We observed a similar pattern in HEK 293T cells transfected with plasmids encoding C-terminally FLAG-tagged full-length TMED7 or its truncated mutants (fig. S7A). Further analysis of the transfected HEK 293T cells with an antibody specific for protein disulfide isomerase (PDI), a marker of the ER, showed that whereas both the CC and GOLD mutants seemed to overlap with PDI, the Delta mutant was distributed more diffusely and did not accumulate in the Golgi (fig. S7B). Therefore, these data suggest that the cytosolic tail of TMED7 is essential for its correct localization within the Golgi, because the loss of the diphenylalanine motif caused the protein to be trapped in the endomembrane system.
Fig. 7. The localization of TMED7 in the Golgi in THP-1 cells depends on its COP II–binding motif.
(A to D) THP-1 cells were transduced with lentiviruses encoding HA-tagged (A) full-length TMED7 or the (B) Delta, (C) CC, or (D) GOLD TMED mutant constructs. Three days later, the cells were permeabilized and the HA-tagged TMED7 constructs were labeled with Alexa Fluor 633 (red), whereas endogenous giantin (a Golgi marker) was labeled with Alexa Fluor 404 (blue). Images are representative of two independent experiments in each of which ~20 cells of each type were observed to have similar localization patterns. Scale bars: 10 μm.
TMED7 facilitates the cell-surface expression of TLR4
The stimulation of TLR4 by LPS can be initiated from either the cell surface or in the Golgi after internalization of LPS (34, 35). Therefore, the cell-surface abundance of TLR4 might determine the extent of LPS responsiveness observed earlier. The N-glycosylation of TLR4 is important for its cell-surface expression and responsiveness to ligand. Studies showed that chaperone molecules are important for this process, as is MD2, which is required for the glycosylation of TLR4 that enables its cell-surface expression (9, 12, 36).
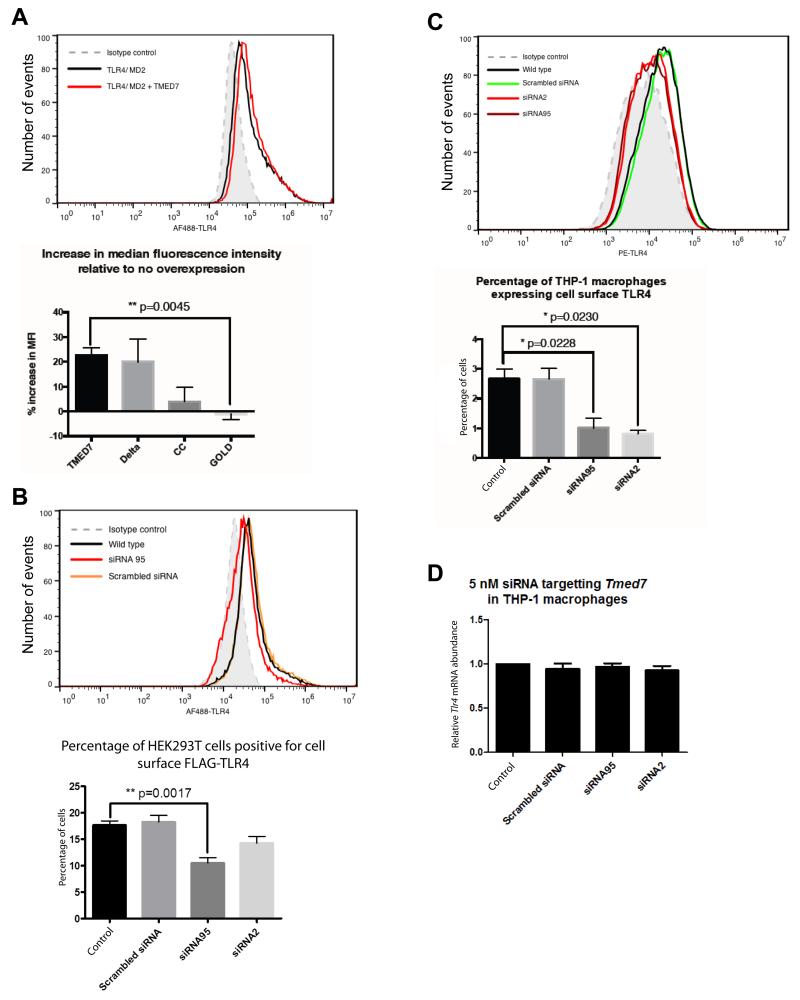
To assess the role of TMED7 in the trafficking of TLR4 to the cell surface, we performed flow cytometric analysis of HEK 293T cells transfected with plasmids encoding FLAG-TLR4 and MD2, as well as of THP-1 cells, which have endogenous TLR4. Because the FLAG tag was located in the N-terminal ectodomain of TLR4 and the cells were not fixed nor permeabilized, only TLR4 molecules present on the surface of HEK 293T cells were observed in the assay. We gated for viable cells based on their forward and side-scatter profiles, and then we analyzed this population for cells that were positive for Alexa Fluor 488 signal (TLR4), with the isotype control set as the signal threshold. Untransfected and unstained cells were included as negative controls to ensure that the signals were not a result of nonspecific antibody binding or auto-fluorescent cells, respectively. We used this method to determine the cell-surface expression of TLR4 under conditions in which TMED7 was reduced by siRNA or increased by overexpression. Transfection of cells with plasmid encoding TMED7 led to an increase in the cell-surface abundance of TLR4 (Fig. 8A). In contrast, knock-down of Tmed7 in HEK 293T cells led to a substantial reduction in the cell-surface expression of FLAG-TLR4 based on the percentage of transfected cells that expressed cell-surface TLR4 (Fig. 8B). We validated these findings by repeating the TMED7 knockdown experiments in THP-1 cells, which showed a decrease in the cell-surface abundance of TLR4 that was not a result of any decrease in the abundance of Tlr4 mRNA (Fig. 8, C and D). Together, these results suggest that TMED7 is involved in the early-stage trafficking of TLR4, which leads to its transport to the cell surface and therefore enhances the signaling activity of the receptor.
Fig. 8. TMED7 enhances the cell-surface abundance of TLR4.
(A) HEK 293T cells were transfected with plasmid encoding FLAG-TLR4 alone or together with plasmids encoding TMED7-HA or HA-tagged TMED7 truncation mutants. Cells were then analyzed by flow cytometry to determine the cell-surface abundance of TLR4. The histogram shows the change in the mean fluorescence intensity (MFI) corresponding to TLR4 upon TMED7-HA overexpression (compare red and black lines), bottom panel represents the increase in median fluorescence intensity (MFI) of cell surface TLR4 staining relative to no overexpression. (B) Knockdown of TMED7 in HEK 293T cells decreases the cell-surface abundance of FLAG-TLR4. HEK 293T cells were transfected with plasmid encoding FLAG-TLR4 alone or in the presence of the indicated siRNAs. 72 hours later, cells were analyzed by flow cytometry with an anti-FLAG antibody. The histogram is from a single representative experiment. Bar graph shows the mean ± SEM from three independent experiments. (C and D) Knockdown of TMED7 in THP-1 cells decreases the cell-surface abundance of endogenous TLR4, but not its total cellular abundance. THP-1 cells were left untreated or were transfected with the indicated siRNAs. 72 hours later, the cells were analyzed by flow cytometry with an anti-TLR4 antibody. The histogram is from a single representative experiment. Bar graph shows the mean ± SEM from three independent experiments. (D) THP-1 cells were left untransfected or were transfected with the indicated siRNAs. Three days later, cells were harvested and the relative amounts of Tlr4 mRNA compared to that of Actb mRNA were determined by qPCR analysis. Data are means ± SEM from three independent experiments. P values were calculated with an unpaired, two-tailed t test with Welch’s correction.
DISCUSSION
We showed that TLR4 is a cargo molecule for TMED7, which facilitates the intracellular trafficking of the receptor before its transport to the cell surface. We presented several lines of evidence that TMED7 is a type I membrane protein with a luminal N-terminal region and a short cytoplasmic domain with a characteristic di-phenylalanine COP II–sorting motif (Fig. 1, A and B). This topology for TMED7 [also known as (g)p27 or p24γ3] is supported by historical studies, including the N-terminal sequencing of native TMED7 isolated from the Golgi membrane fractions of rat, which revealed a cleaved signal peptide (19). Our conclusions are not, however, consistent with a study that reported that TMED7 is anchored in the membrane by the N-terminal helix and that the GOLD domain is exposed to the cytoplasm (32). This conclusion was drawn from membrane fractionation assays by mutating each of the helical regions individually, and by protease protection assays, although the experiment presented lacks a positive control to show that a bona fide ER lumen protein is protected from degradation under the conditions used. Doyle et al. also concluded that TMED7 interacts directly with the signaling adaptor TRAM and a TMED7-TRAM splice hybrid [TRAM with GOLD domain (TAG)] (37), both of which acted as negative regulators of TLR4 signaling from the endosome. In contrast, we did not detect a direct interaction between TMED7 and TRAM in unstimulated cells, a result that is consistent with the membrane topology and function that we propose. In the case of the TAG construct, a splicing product that includes the GOLD and coiled-coil domains of TMED7 joined to the TIR domain of TRAM, it is possible be that it is secreted and has a dominant-negative effect by forming heterodimers with endogenous TMED7 in the lumen of the ER. This question will be addressed further in the future by cell fractionation and immune-EM approaches.
We showed that the luminal coiled-coil and GOLD domains of TMED7 were sufficient to mediate binding to TLR4; therefore, it is likely the ectodomain of TLR4 that mediates binding to TMED7. The GOLD domain alone does not form dimers and cannot bind to TLR4, which suggests that recognition requires the homodimerization of TMED7 or its heterodimerization with other TMED family members (24). This result also establishes, as expected, that the domain predicted to form an α-helical, coiled-coil mediates dimerization. This is an important feature, because TMED7 must associate with TMED2, TMED9, and TMED10 to be exported from the ER to the Golgi and for its subsequent dynamic recycling within the early secretory pathway (24).
Although TMED7 forms homodimers (26), we cannot rule out the possibility that heterodimers containing TMED7 interact with TLR4. Whereas TMED2 and TMED7 contain only COP II–sorting signals, TMED9 and TMED10 contain both COP I– and COP II–sorting signals (Phe-Phe and Lys-Lys) in their cytosolic tails. Previous studies showed that KK-to-SS mutations in the cytosolic tails of TMED9 and TMED10 result in the redistribution of TMED2 and TMED7, as well as that of TMED9 and TMED10, to the cell surface (19, 25). This is presumably a result of a defect in the retrograde transport of TMED proteins through COP I–containing vesicles back to the ER. Thus, we have two distinct groups of p24 proteins: one, including TMED2 and TMED7, whose sole function is to mediate anterograde trafficking, and the other, including TMED9 and TMED10, whose role is to retrieve ER-resident proteins back from post-ER compartments, as well as to mediate anterograde transport. In future studies, we will co-express these p24 proteins together with TLR4 to determine whether hetero-dimerization is required for TLR4 recognition or TMED7 recycling back to the ER, or both. The TMED7 Delta and CC truncation mutants, which lack the anterograde sorting signal for COP II, also bound to TLR4, but did not accumulate in the Golgi. These data suggest that it is likely that trafficking of TLR4 to the Golgi requires segregation into COP II–coated vesicles by homodimers or heterodimers of TMED7, and that the association of TMED7 and TLR4 in a stable complex defines receptors that are transiting the early secretory pathway.
The presence of the adaptor protein TRAM caused substantially enhanced the binding of TMED7 to TLR4. This finding could indicate that TRAM becomes pre-associated with the TIR domain of TLR4 during biosynthesis, and that this interaction inhibits recognition of the COP II–sorting motif and anterograde trafficking, leading to an accumulation of TMED7 and TLR4 in the ER. In this regard, previous studies showed that TLR4 and TRAM co-localize in the Golgi and at the cell surface, which is suggestive of coordinated trafficking of the receptor and the adaptor (38). TRAM is modified by the addition of the fatty acid myristate to an N-terminal glycine residue. The myristoyl group is added by the enzyme N-myristoyl transferase, and the modified proteins are cotranslationally inserted into the ER membrane (39). Thus, another explanation for the observed accumulation of TMED7-TLR4 complexes might be that the increased abundance of TRAM sterically blocks the formation of the COP II coats that are required for anterograde transport, again inhibiting transport from the ER to the Golgi. The interaction between TMED7 and TLR4 seemed to be weakened by co-expression of CD14 and MD2, individually and together, which was not a result of the varying total abundances of these proteins (Fig. 5C). To date, there has been no evidence that TLR4 and CD14 form a direct association with each other. Most studies of CD14 in the context of TLR4 signaling have been focused on its role in the transfer of LPS to the TLR4-MD2 complex on the cell surface and its subsequent endocytosis to early endosomes where TRIF dependent signaling is initiated (4, 35, 40, 41). One possibility is that the presence of CD14 enhances the endocytosis of TLR4 and that this is coupled with increased trafficking from the Golgi to the cell surface. This step in secretion probably involves the dissociation of TMED7 from the nascent receptor.
The TMED7 Delta truncation mutant enhanced constitutive signaling by TLR4, which led to a 20-fold increase in the activation of the IFN-β and NF-κB reporters, suggesting that proper regulation of early-stage secretion is necessary to repress any ligand-independent signaling that could produce potentially dangerous inflammatory responses. Expression of wild-type TMED7 and the TMED7 CC truncation mutant, but not the TMED7 GOLD mutant, led to a lower increase in TLR4 constitutive activity of about 2- to 5-fold. Constitutive activation of TLR4 can occur when the receptors accumulate in membranes at high abundance. The ectodomain of TLR4 acts as a constitutive suppressor of TLR4 activation, and this repression can be relieved by exposure to ligand, overexpression, truncation, or substitution with domains that have a propensity for dimerization (42). In the case of cells expressing the TMED7 CC mutant, the constitutive signal may originate from intracellular compartments, because trafficking of TLR4 through the Golgi appeared to be blocked in these cells, leading to intracellular accumulation of the receptor. On the other hand, the increased TLR4 activity caused by overexpression of full-length TMED7 is probably the result of enhanced trafficking of TLR4 to the cell surface (Fig. 8). The TMED7 Delta mutant enhanced TLR4 signaling in the presence and absence of LPS, which suggests that the intracellular accumulation of TLR4 stimulates its LPS-independent activation. Overall, these results suggest a role for the di-phenylalanine COP II–sorting motif in the cytosolic tail of TMED7, as well as for the residues Glu193 and Gln204 in its transmembrane domain, in TLR4 trafficking (17, 19, 33). These studies showed that interactions between these key residues affect not only the localization of TMED7, but also the rate of the anterograde trafficking of the TMED protein in a regulated manner (33).
We found that whereas overexpression of full-length TMED7 affected both MyD88 and TRIF/TRAM-dependent signaling pathways, knockdown of TMED7 affected only MyD88-dependent TLR4 signaling. It is possible that this selectivity occurs because these two pathways are initiated from two distinct locations: the cell surface and early endosomes (2, 5). Kagan et al. proposed the sequential initiation of MyD88-dependent signaling at the cell surface, which is followed by endocytosis of TLR4 and the initiation of TRAM-dependent signaling in the early endosomes. The model put forward by Husebye et al. suggests that there are two distinct TLR4 populations on the cell surface and in the endocytic recycling compartment (ERC) of human monocytes that can respond to infection with Escherichia coli independently of each other. From these conclusions, we can postulate that whereas initiation of the MyD88-dependent pathway upon TLR4 stimulation relies on the transport of the receptor to the cell surface, the activation of the TRAM-dependent pathway relies on both the endocytosis of the activated receptor and the trafficking of ligand-free TLR4 in the ERC to the ligand-containing phagosome, a process that is Rab11a-dependent. Therefore, we propose that the main role of TMED7 is to traffic TLR4 to the cell surface and not to the other intracellular compartments, such as the ERC. This would explain why deletion of TMED7 affected only the MyD88-dependent signaling pathway.
Little is known about the molecular mechanisms by which TMED proteins associate with their cargoes in the ER; however, the available evidence suggests that the secretion of cargo proteins is coupled to N-glycosylation in the ER. For example, a systematic mutagenesis of potential N-glycosylation sites in TLR2 showed that all four are modified and contribute to efficient trafficking of the receptor to the plasma membrane, although a conserved site at the C-terminus is of particular importance (43). In the case of TLR4, there are nine residues that are N-glycosylated, and two of these, Asn526 and Asn575, are essential for trafficking to the membrane, whereas other sites have a more partial effect on the cell-surface expression of the receptor (12). Thus, an attractive hypothesis is that TMED7, acting as a quality control step, assembles with TLR4 by recognizing the nascent glycosylated receptor in the ER lumen and marks the receptor as cargo for anterograde transport in COP II–containing vesicles to ERGIC and Golgi compartments.
Another aspect of interest is the mechanism by which TLR4 is released from TMED7 in the Golgi. Rab10 promotes the trafficking of TLR4 from the Golgi to the cell surface (15). It is therefore possible that TMED7 and Rab10 work together in the process of transporting TLR4 to the cell surface. However, the precise molecular events that lead to this have not yet been elucidated. The acidic pH of the compartment in the late secretory pathway could lead to the dissociation of TMED from its cargo (44); a similar general mechanism also operates to cause the release of low-density lipoprotein (LDL) from its receptor after internalization into acidic endosomes (45). A more tightly regulated switch could be implemented, such as that used by TMED2 in the resensitization of protease-activated receptor 2 (PAR2) on the cell surface from the Golgi (46). In the resting state, TMED2 associates with its PAR2 cargo in the Golgi. Upon stimulation of PAR2 on the cell surface, the receptors are endocytosed, and the cell surface has to be replenished with receptor proteins, an event that is mediated by the activation of a small GTP-binding protein ADP-ribosylation factor 1 (ARF1), which leads to the dissociation of PAR2 from TMED2. This mechanism might explain the decrease in the amount of TMED7 associated with the TLR4 upon stimulation of HEK 293T cells with LPS (Fig. 2D). Perhaps a similar mechanism is used in this particular system to resensitize TLR4 molecules? In future studies, we aim to explore the nature of the interactions between TMED dimers and glycans of the TLR ectodomain at a molecular level, and to test the hypothesis that disassembly of TLR4 in the Golgi is mediated by the transition to low pH and the Golgi-specific modification of the glycans.
MATERIALS AND METHODS
Constructs and antibodies
Complementary DNAs (cDNAs) encoding human TLR4 and human CD14 in pcDNA3, human MD2 in pEFIRES, and p125-luc were generously provided by Dr Clare Bryant, University of Cambridge, UK. The plasmid p125-luc contains the promoter region of IFNB1 gene followed by firefly luciferase as described previously (47). The plasmids phRG-TK and pNF-κB-luc were purchased from Promega and Clontech, respectively. A pEF-BOS plasmid encoding TRAM with C-terminal FLAG and His tags (TRAM-FLAG) and a pcDNA3 plasmid encoding TLR4 with N-terminal FLAG tag (FLAG-TLR4) were gifts from Dr Ashley Mansell, MIMR, Australia. The plasmid TMED7-GFP in EX-V1709-M03 (OmicsLink) was a gift from Prof. Luke O’Neill, Trinity College Dublin, Ireland. The plasmids pHR:TLR4Cit and pHR:TLR3Cit, which encode C-terminal mCitrine tags, were gifts from Dr Brett Verstak. Primary antibodies used in this study include rabbit and chicken anti-HA antibodies (ab9110 and ab9111 respectively, from Abcam), mouse anti-FLAG antibody (Sigma, F3165), immunoglobulin G1 (IgG1) isotype control antibody from murine myeloma (Sigma, M5284), rabbit anti-giantin antibody (abcam, ab24586), mouse anti-PDI antibody (abcam, ab5484), which recognizes the ER marker RL77, phycoerythrin (PE)-conjugated anti-human CD284 (TLR4, eBioscience, 12-9917), and PE-conjugated mouse IgG2a K isotype control antibody (eBioscience, 12-4724). Secondary antibodies used in this study were: goat anti-rabbit IgG (whole molecule)-peroxidase (Sigma, A0545), goat anti-mouse IgG (whole molecule)-peroxidase (Sigma, A4416), and Mouse TrueBlot ULTRA: anti-mouse Ig HRP (eBioscience, 18-8817) for Western blotting. For immunofluorescence analysis, we used Alexa Fluor 633–conjugated goat anti-chicken IgG (H+L) (Molecular Probes, A21103), Alexa Fluor 405–conjugated goat anti-rabbit IgG (H+L) (Molecular Probes, A31556), and Alexa Fluor 488–conjugated goat anti-mouse IgG (H+L) (Molecular Probes, A11001).
Cloning and mutagenesis
Mammalian expression vectors encoding human TMED7 and its truncated mutants were constructed with TMED7-GFP in EX-V1709-M03 as the template with the Gateway cloning technology (Invitrogen). Standard PCR assays were performed with Vent polymerase (NEB). The constructs generated were called TMED7 (full-length, residues 1 to 224), CC (residues 1 to 187), and GOLD (residues 1 to 144). The attB1 and attB2 sites were incorporated into the forward and reverse primers, respectively, flanking the cDNA, to clone the cDNA into the pDONR221 entry vector by Gateway BP reaction with BP clonase enzyme. One forward primer contained an attB1 site adjacent to the native signal peptide of TMED7 in the 5′ to 3′ direction. Three reverse primers, for each of the three constructs, consisted of an attB2 site, stop codon, FLAG or HA tag, and then the cDNA encoding the TMED7 construct in the 5′ to 3′ direction. The Gateway LR reaction was then performed with the enzyme LR clonase to insert the cDNA into a Gateway pcDNA-DEST47 destination vector, which could be used to transfect mammalian cells. The cDNA needed to encode another mutant TMED7, Delta (corresponding to residues 1 to 210, lacking the cytoplasmic tail) with a C-terminal HA tag, was generated with the Quikchange mutagenesis kit with PfuTurbo (Stratagene). Primer pairs for mutagenesis were designed to skip the cytoplasmic tail to be deleted, and cDNA encoding TMED7-HA was used as the template in the PCR reaction. Primer sequences are available in tables S1 and S2. For lentivirus production, the cDNAs of interest were cloned into the pHR vector. Briefly, PCR reactions were performed to amplify the cDNAs of interest and to incorporate Bam HI and Not I sites at the 5′ and 3′ ends of the cDNAs, respectively. The PCR products and pHR:TLR4Cit were digested with Bam HI and Not I (NEB, R3136 and R3189) according to the manufacturer’s instructions and then were purified. Ligation reactions were performed with T4 DNA Ligase (NEB, M202).
Cell culture and transfections
HEK 293 and HEK 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma, D5546) supplemented with 2 mM L-glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% (v/v) heat-inactivated fetal calf serum (FCS) at 37°C, 5% CO2. The jetPEI transfection reagent (Polyplus transfection) was used to transfect cells with DNA plasmids according to the manufacturer’s protocol. Cells in each well of a 96-well plate were transfected with a total of 100 ng of DNA plasmid. Cells grown in 6-well plates were transfected with a total of 3 μg of DNA plasmid per well. For detailed accounts of the amount of DNA used for different assays, see tables S3 to S5. Cells were analyzed 24 to 48 hours after transfection. THP-1 cells were obtained from Dr Clare Bryant. The cells were maintained in suspension in RPMI-1640 with L-glutamine and sodium bicarbonate (Sigma, R8758), supplemented with 10% FCS, penicillin (100 units/ml), streptomycin (100 μg/ml), 25 mM HEPES, and 20 μM β-mercaptoethanol. Before being transfected with siRNAs, THP-1 cells were first differentiated into macrophage-like cells. For differentiation, cells were treated with phorbol myristate acetate (PMA, 100 ng/ml, Sigma, P8139) in complete RPMI for two days before subsequent analysis.
Lentivirus production
HEK 293T cells were transfected with the plasmids pMDG (which encodes the VSV-G envelope) and pΔ8.91 (for packaging) together with individual pHR plasmids expressing genes of interest for lentivirus production. Cells (5 × 105) were seeded in 10-cm cell culture dishes in 10 ml of complete DMEM two days before transfection. For one dish, HEK 293T cells were transfected with 7.2 μg of pΔ8.91 and pHR and 3.6 μg of pMDG. For one transfection, DNA plasmids were mixed in 150 mM NaCl to give a total volume of 250 μl. In a separate tube, 30 μl of jetPEI transfection reagent (Polyplus transfection) were mixed with 220 μl of 150 mM NaCl. The jetPEI mix was added dropwise onto the DNA mix and incubated at room temperature for 30 min. After incubation, the DNA-jetPEI mix was added to the cells. The following day, the culture medium was replaced with antibiotic-free DMEM supplemented with 2 mM L-glutamine and 10% FCS, and the cells were incubated for two more days before lentivirus was harvested. THP-1 cells were incubated with 1 ml of lentivirus per well in a 6-well plate. The following day, the culture medium was replaced with 2 ml of complete RPMI-1640, and cells were incubated for three to four days before experiments were performed.
Recombinant protein expression in insect cells
To generate recombinant TMED7 CC mutant protein, the cDNA encoding amino acid residues 35 to 187 of TMED7 containing the endogenous signal peptide (SP-CC) or a pre-pro-trypsin leader sequence (PP-CC) was generated with 5′ Bam HI and a 3′ Age I restriction sites. The inserts were introduced into the pFastBac-1 transposition vector (Invitrogen), which encodes a C-terminal Fc domain of human IgG1 and 6 × His tags for purification and detection purposes, as described previously (48, 49). The recombinant proteins were produced in a baculovirus expression system by infecting Trichoplusia ni insect cells with the baculoviruses generated, as previously described (49). Two days after infection, the secreted recombinant protein in the supernatant was purified with a HiTrap rProtein A FF 5-ml column (GE Healthcare, 17-5080). Purified proteins were resolved on 10% SDS PAGE gels and were transferred to polyvinylidene difluoride membranes and sent for N-terminal sequencing at the departmental protein and nucleic acid analysis facility. The first five to six amino acid residues were identified.
siRNAs, RNA extraction, and qPCR analysis
TMED7-specific siRNA duplexes (HSS121495, or siRNA95 for short) were purchased from Invitrogen (Stealth siRNA) and from Sigma (siRNA2 and siRNA3). The siRNA2 and siRNA3 duplexes were designed in our lab with the siRNA target finder tool from Ambion. Generated sequences were subsequently ranked using parameters described previously (50), and only those two sequences with the highest scores were selected. The sequences of the siRNA duplexes are shown in table S6. HEK 293 and HEK 293T cells in 6-well dishes were transfected with siRNA with the jetPRIME transfection reagent (Polyplus transfection) and incubated for three days, after which they were transfected with plasmid DNAs or were used for data collection. Transfection of macrophage-like THP-1 cells with siRNA was performed with the INTERFERin (Polyplus transfection) two days after the cells were treated with PMA, and the cells were incubated for a further three days before being analyzed by ELISA or flow cytometry. Total RNA from cells was purified with the RNeasy Plus kit and QiaShredder (QIAgen). First-strand cDNA was synthesized from the total RNA with SuperScript II reverse transcriptase (Invitrogen) and random hexamers (Promega, C1181) as described in the protocol from Untegasser lab (http://www.untergasser.com/lab/protocols/cdna_synthesis_superscript_ii_v1_0.htm). The cDNA served as template for real-time quantitative PCR (qPCR) analysis to determine the efficiency of gene knockdown with Platinum SYBR Green qPCR SuperMix (Invitrogen, 11733038). The relative abundance of TMED7 mRNA was determined and normalized to that of the internal control, GAPDH (for HEK 293T cells) or β-actin (for THP-1 cells) to determine the relative extent of target knockdown with the Livak method of 2(-ΔΔCt)(51). Primers specific for TMED7, TRAM, TLR4, β-actin, and GAPDH were designed to span exon-exon junctions to eliminate the amplification of genomic DNA (see table S7).
Luciferase reporter assays
The protocol used was adapted from a previously described method (52). Briefly, HEK 293 cells were transfected with the appropriate expression plasmids with the jetpEI transfection reagent. The reporter genes used were p125-luc and pNF-κB-luc. For knockdown experiments, cells were transfected with siRNA three days before being transfected with plasmid DNA, as described earlier. Forty-eight hours after, cells were stimulated for 6 hours with LPS from E. coli O127:B8 (Sigma Aldrich, L3129), poly I:C HMW (Invivogen, tlrl-pic), or flagellin from S. typhimurium (Invivogen, tlrl-stfla) diluted in serum-free DMEM. The cells were washed in phosphate-buffered saline (PBS) and then lysed with passive lysis buffer (Promega) diluted in PBS. Firefly and Renilla luciferase activities were quantified individually with luciferin and coelenterazine reagents, respectively, with a LUMIstar Omega luminometer (BMG Labtech).
Enzyme-linked immunosorbent assay
Three days after THP-1 cells were transfected with siRNAs, the cells were stimulated for 24 hours with LPS diluted in serum-free RPMI-1640 at the concentrations indicated in the figures. The amounts of human IL-6, TNF-α, and RANTES produced by the cells were measured in the culture supernatant according to the protocols from the ELISA kits (R&D Systems).
Co-immunoprecipitations and Western blotting analysis
Forty-eight to 72 hours after transfection or lentiviral transduction, cells were lysed in cold lysis buffer [50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EDTA, 2 mM MgCl2, 2 mM CaCl2 containing 1% Triton X-100, 0.5% n-Octylglucoside, and supplemented with 1 mM PMSF and 1x protease inhibitor cocktail set V (Calbiochem, 535141)]. Soluble extracts of cell lysates were collected by centrifugation at 15000g for 10 min. FLAG-tagged recombinant proteins in the supernatants were immunoprecipitated by incubation with anti-FLAG M2 magnetic beads (Sigma, M8823) with a DynaMag-2 magnet (Invitrogen) to separate beads from wash buffer. Total protein and co-immunoprecipitated samples were analyzed by Western blotting with the antibodies indicated in the figures.
Enzymatic deglycosylation
Proteins were harvested from HEK 293T cells in lysis buffer as described earlier, but with 1% NP-40 used as the detergent. TMED7-HA from the cell lysates was treated overnight at 37°C with 1000 U PNGaseF (NEB) according to the manufacturer’s protocol, or with enzyme-free buffers as a negative control. SDS loading buffer containing reducing agent was added to the beads and boiled at 100°C for 10 min. Samples were then analyzed by Western blotting.
Confocal microscopy
Cells were seeded on coverslips in 6-well plates. Two to three days after transfection or transduction of the cells, culture medium was aspirated from the wells, and the cells on coverslips were washed once in cold PBS. The cells were then fixed with 4% paraformaldehyde (PFA) in PBS and permeabilized with 0.2% Triton x-100 in PBS at room temperature. Cells were washed twice between steps in cold PBS. For immunohistochemistry, cells were blocked in blocking buffer [5% bovine serum albumin (BSA) in PBS with 0.1% Tween-20 (PBST)] for 30 min at room temperature. Cells were stained in a cocktail of primary chicken antibodies against HA and rabbit antibodies against giantin (all diluted 1:200 in blocking buffer) overnight in the fridge. Cells were then washed three times in PBST before being incubated in a cocktail of Alexa Fluor–conjugated secondary antibodies against chicken (AF633) and rabbit (AF405) antibodies (all diluted 1:200 in blocking buffer) for 1 hour at room temperature. Coverslips were then inversely mounted onto glass SuperFrost Plus 76 × 26mm glass slides (VWR International) containing two drops of Vectashield hardset mounting medium (Vectorlabs). A Leica SP5 Upright (laser wavelengths used: 405 nm, 514 nm, and 633 nm) was used to visualize the cells. Leica LAS AF software was used to obtain the fluorescent micrographs.
Flow cytometric analysis
Two days after transfection, HEK 293T cells were detached from the bottoms of the wells with accutase (PAA), washed once in PBS, and blocked in blocking buffer (5% BSA in PBS) for 30 min at room temperature. Cells were stained with anti-FLAG antibody (10 μg/ml) or mouse IgG1 isotype control in blocking buffer overnight in the fridge, washed, and then incubated with Alexa Fluor 488–conjugated secondary antibody for 1 hour at room temperature in the dark. Cells were washed and analyzed with a BD Accuri C6 flow cytometer. Three days after they were transfected with siRNAs, macrophage-like THP-1 cells were detached from the bottoms of the wells with cold 10 mM EDTA in PBS, washed once in 0.5% BSA in PBS, and blocked in blocking buffer for 30 min at room temperature. Cells were stained with PE-conjugated anti-CD284 (TLR4) antibody (2 μg per test) or PE-conjugated mouse IgG2a K isotype control antibody (0.5 μg per test) in blocking buffer overnight at 4°C. Cells were washed twice and analyzed with BD Accuri C6 flow cytometer.
Statistical analysis
All graphs were generated with Prism5 software (GraphPad), and statistical analyses were performed with unpaired, two-tailed t tests with Welch’s correction to obtain P values where appropriate.
Online prediction tools for protein analysis
Clustal Omega (EMBL-EBI) was used to generate multiple amino acid sequence alignments. SignalP 4.0 and NetNGlyc (CBS, University of Denmark) were used to predict the signal peptide and N-glycosylation site in a secreted protein. TMHMM (CBS, University of Denmark) prediction was performed to identify the transmembrane domain of a protein and its membrane topology. Psipred (University College, London) was used to predict the secondary structure of a protein.
Supplementary Material
Acknowledgments
We thank B. Verstak for his assistance in lentivirus production, and J. Sakai for his help with setting up the ELISA assays. Funding: This work was supported by programme grants from the Wellcome Trust (WT081744/Z/06/Z) and the UK Medical Research Council (G1000133) to N.J.G and CEB and Wellcome Investigator award to NJG (WT100321/z/12/Z). We thank Cath Green and Matt Wyland for confocal miscroscopy.
Footnotes
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Bryant CE, Spring DR, Gangloff M, Gay NJ. The molecular basis of the host response to lipopolysaccharide. Nat. Rev. Microbiol. 2010;8:8–14. doi: 10.1038/nrmicro2266. [DOI] [PubMed] [Google Scholar]
- 2.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 4.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas O, Nilsen NJ, Stenmark H, Latz E, Lien E, Mollnes TE, Bakke O, Espevik T. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husebye H, Halaas O, Stenmark H, Tunheim G, Sandanger O, Bogen B, Brech A, Latz E, Espevik T. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 2006;25:683–692. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Shibata T, Akashi-Takamura S, Kiyokawa T, Wakabayashi Y, Tanimura N, Kobayashi T, Matsumoto F, Fukui R, Kouro T, Nagai Y, Takatsu K, Saitoh S, Miyake K. A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J. Exp. Med. 2007;204:2963–2976. doi: 10.1084/jem.20071132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi Y, Kobayashi M, Akashi-Takamura S, Tanimura N, Konno K, Takahashi K, Ishii T, Mizutani T, Iba H, Kouro T, Takaki S, Takatsu K, Oda Y, Ishihama Y, Saitoh S, Miyake K. A protein associated with toll-like receptor 4 (PRAT4A) regulates cell surface expression of TLR4. J. Immunol. 2006;177:1772–1779. doi: 10.4049/jimmunol.177.3.1772. [DOI] [PubMed] [Google Scholar]
- 10.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell. Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 11.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 12.da Silva Correia J, Ulevitch RJ. MD-2 and TLR4 N-linked glycosylations are important for a functional lipopolysaccharide receptor. J. Biol. Chem. 2002;277:1845–1854. doi: 10.1074/jbc.M109910200. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi T, Muroi M, Tanamoto K. MD-2 is necessary for the toll-like receptor 4 protein to undergo glycosylation essential for its translocation to the cell surface. Clin. Diagn. Lab. Immunol. 2003;10:405–410. doi: 10.1128/CDLI.10.3.405-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gioannini TL, Teghanemt A, Zhang DS, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc. Natl. Acad. Sci. U S A. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Lou J, Ouyang C, Chen W, Liu Y, Liu X, Cao X, Wang J, Lu L. Ras-related protein Rab10 facilitates TLR4 signaling by promoting replenishment of TLR4 onto the plasma membrane. Proc. Natl. Acad. Sci. U S A. 2010;107:13806–13811. doi: 10.1073/pnas.1009428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nature Rev. Mol. Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- 18.Kappeler F, Klopfenstein DR, Foguet M, Paccaud JP, Hauri HP. The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J. Biol. Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez M, Dejgaard K, Fullekrug J, Dahan S, Fazel A, Paccaud JP, Thomas DY, Bergeron JJM, Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dancourt J, Barlowe C. Protein Sorting Receptors in the Early Secretory Pathway. Annu. Rev. Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 21.Strating JR, van Bakel NH, Leunissen JA, Martens GJ. A comprehensive overview of the vertebrate p24 family: identification of a novel tissue-specifically expressed member. Mol. Biol. Evol. 2009;26:1707–1714. doi: 10.1093/molbev/msp099. [DOI] [PubMed] [Google Scholar]
- 22.Strating JR, Martens GJ. The p24 family and selective transport processes at the ER Golgi interface. Biol. Cell. 2009;101:495–509. doi: 10.1042/BC20080233. [DOI] [PubMed] [Google Scholar]
- 23.Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms JB, Wieland FT. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J. Cell. Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fullekrug J, Suganuma T, Tang BL, Hong WJ, Storrie B, Nilsson T. Localization and recycling of gp27 (hp24 gamma(3)): Complex formation with other p24 family members. Mol. Biol. Cell. 1999;10:1939–1955. doi: 10.1091/mbc.10.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emery G, Rojo M, Gruenberg J. Coupled transport of p24 family members. J. Cell Sci. 2000;113:2507–2516. doi: 10.1242/jcs.113.13.2507. [DOI] [PubMed] [Google Scholar]
- 26.Jenne N, Frey K, Brugger B, Wieland FT. Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 2002;277:46504–46511. doi: 10.1074/jbc.M206989200. [DOI] [PubMed] [Google Scholar]
- 27.Stamnes MA, Craighead MW, Hoe MH, Lampen N, Geromanos S, Tempst P, Rothman JE. An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc. Natl. Acad. Sci. U S A. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belden WJ, Barlowe C. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell. 2001;12:957–969. doi: 10.1091/mbc.12.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denzel A, Otto F, Girod A, Pepperkok R, Watson R, Rosewell I, Bergeron JJM, Solari RCE, Owen MJ. The p24 family member p23 is required for early embryonic development. Curr. Biol. 2000;10:55–58. doi: 10.1016/s0960-9822(99)00266-3. [DOI] [PubMed] [Google Scholar]
- 31.Jerome-Majewska LA, Achkar T, Luo L, Lupu F, Lacy E. The trafficking protein Tmed2/p24beta(1) is required for morphogenesis of the mouse embryo and placenta. Dev. Biol. 2010;341:154–166. doi: 10.1016/j.ydbio.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle SL, Husebye H, Connolly DJ, Espevik T, O'Neill LAJ, McGettrick AF. The GOLD domain-containing protein TMED7 inhibits TLR4 signalling from the endosome upon LPS stimulation. Nat. Commun. 2012;3:707–714. doi: 10.1038/ncomms1706. [DOI] [PubMed] [Google Scholar]
- 33.Fiedler K, Rothman JE. Sorting determinants in the transmembrane domain of p24 proteins. J. Biol. Chem. 1997;272:24739–24742. doi: 10.1074/jbc.272.40.24739. [DOI] [PubMed] [Google Scholar]
- 34.Hornef MW, Normark BH, Vandewalle A, Normark S. Intracellular recognition of lipopolysaccharide by toll-like receptor 4 in intestinal epithelial cells. J. Exp. Med. 2003;198:1225–1235. doi: 10.1084/jem.20022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J. Biol. Chem. 2002;277:47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 36.Rocuts F, Ma Y, Zhang X, Gao W, Yue Y, Vartanian T, Wang H. Carbon monoxide suppresses membrane expression of TLR4 via myeloid differentiation factor-2 in betaTC3 cells. J. Immunol. 2010;185:2134–2139. doi: 10.4049/jimmunol.0902782. [DOI] [PubMed] [Google Scholar]
- 37.Palsson-McDermott EM, Doyle SL, McGettrick AF, Hardy M, Husebye H, Banahan K, Gong M, Golenbock D, Espevik T, O'Neill LA. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat. Immunol. 2009;10:579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- 38.Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, Yamamoto M, Akira S, O'Neill L A, Fitzgerald KA, Golenbock DT. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc. Natl. Acad. Sci. U S A. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 40.Akashi S, Saitoh S, Wakabayashi Y, Kikuchi T, Takamura N, Nagai Y, Kusumoto Y, Fukase K, Kusumoto S, Adachi Y, Kosugi A, Miyake K. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: higher affinity than that with MD-2 or CD14. J. Exp. Med. 2003;198:1035–1042. doi: 10.1084/jem.20031076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex. transfer from CD14 to TLR4 and MD-2. J. Biol. Chem. 2001;276:21129–35. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 42.Panter G, Jerala R. The ectodomain of the Toll-like receptor 4 prevents constitutive receptor activation. J. Biol. Chem. 2011;286:23334–23344. doi: 10.1074/jbc.M110.205419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber AN, Morse MA, Gay NJ. Four N-linked glycosylation sites in human toll like receptor 2 cooperate to direct efficient biosynthesis and secretion. J. Biol. Chem. 2004;279:34589–34594. doi: 10.1074/jbc.M403830200. [DOI] [PubMed] [Google Scholar]
- 44.Fujita M, Watanabe R, Jaensch N, Romanova-Michaelides M, Satoh T, Kato M, Riezman H, Yamaguchi Y, Maeda Y, Kinoshita T. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 2011;194:61–75. doi: 10.1083/jcb.201012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 46.Luo W, Wang Y, Reiser G. p24A, a type I transmembrane protein, controls ARF1-dependent resensitization of protease-activated receptor-2 by influence on receptor trafficking. J. Biol. Chem. 2007;282:30246–30255. doi: 10.1074/jbc.M703205200. [DOI] [PubMed] [Google Scholar]
- 47.Yoneyama M, Suhara W, Fukuhara Y, Sato M, Ozato K, Fujita T. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3) J. Biochem. 1996;120:160–169. doi: 10.1093/oxfordjournals.jbchem.a021379. [DOI] [PubMed] [Google Scholar]
- 48.Lewis M, Arnot CJ, Beeston H, McCoy A, Ashcroft AE, Gay NJ, Gangloff M. Cytokine Spatzle binds to the Drosophila immunoreceptor Toll with a neurotrophin-like specificity and couples receptor activation. Proc. Natl. Acad. Sci. U S A. 2013;110:20461–20466. doi: 10.1073/pnas.1317002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber AN, Gangloff M, Moncrieffe MC, Hyvert Y, Imler JL, Gay NJ. Role of the Spatzle Pro-domain in the generation of an active toll receptor ligand. J. Biol. Chem. 2007;282:13522–13531. doi: 10.1074/jbc.M700068200. [DOI] [PubMed] [Google Scholar]
- 50.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Walsh C, Gangloff M, Monie T, Smyth T, Wei B, McKinley TJ, Maskell D, Gay N, Bryant C. Elucidation of the MD-2/TLR4 interface required for signaling by lipid IVa. J. Immunol. 2008;181:1245–1254. doi: 10.4049/jimmunol.181.2.1245. [DOI] [PubMed] [Google Scholar]
- 53.Emery G, Gruenberg J, Rojo M. The p24 family of transmembrane proteins at the interface between endoplasmic reticulum and Golgi apparatus. Protoplasma. 1999;207:24–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.