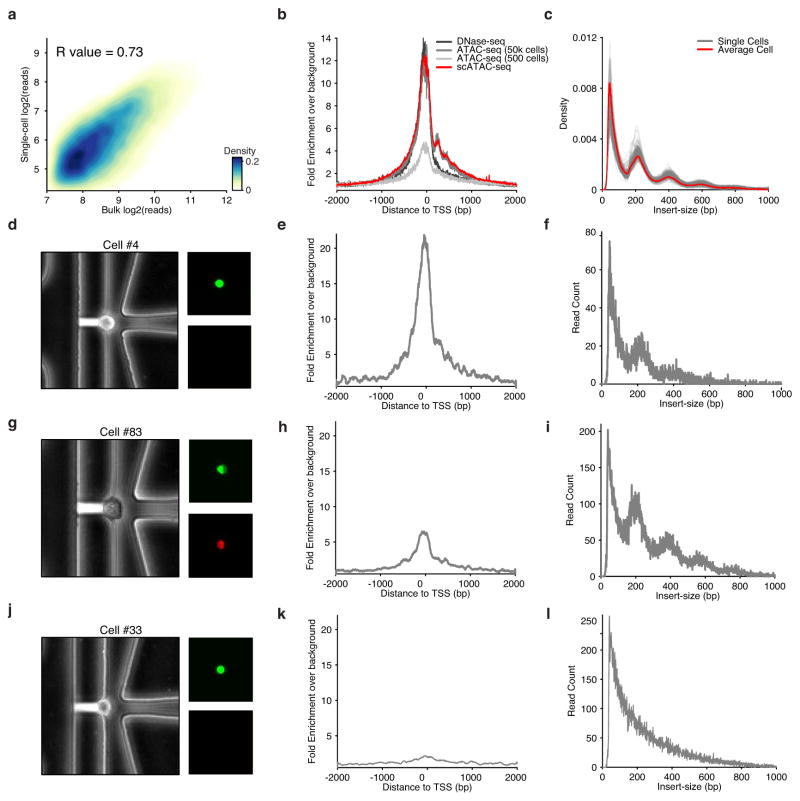
Extended Data Figure 2. scATAC-seq data recapitulate bulk ATAC-seq characteristics.
(a) Reads observed in open chromatin peaks identified from aggregate scATAC-seq data (N = 384 libraries) are highly correlated with reads observed from bulk ATAC-seq. (b) Histogram of aggregated read starts around all TSSs (in K562 cells) comparing ensemble approaches, including 500 cell ATAC-seq reported in a previous publication, to scATAC-seq shows high enrichment above background level of reads. (c) DNA fragment size distribution of ATAC-seq fragments from single cells (grey) and the average of all single cells (red) display characteristic nucleosome-associated periodicity. (d) Phase-contrast (left) and epifluorescence images (right) of captured cell #4 displaying characteristic live cell stain (Calcein) and exclusion of EtBr. (e) Histogram of read starts around TSSs for cell #4 shows high enrichment. (f) DNA fragment size distribution for cell #4 showing nucleosomal periodicity. (g) Images similar to (d) showing staining of cell #83, suggesting low viability due to EtBr staining. (h) Histogram of read starts around TSSs shows lower enrichment than cell #4. (i) DNA fragment size distribution for cell #83. (j) Images similar to (d) showing staining of cell #33 suggesting viability. (k) Histogram of read starts around TSSs of this cell shows low levels of enrichment. (l) DNA fragment size distribution showing no nucleosome-associated periodicity.