Figure 3.
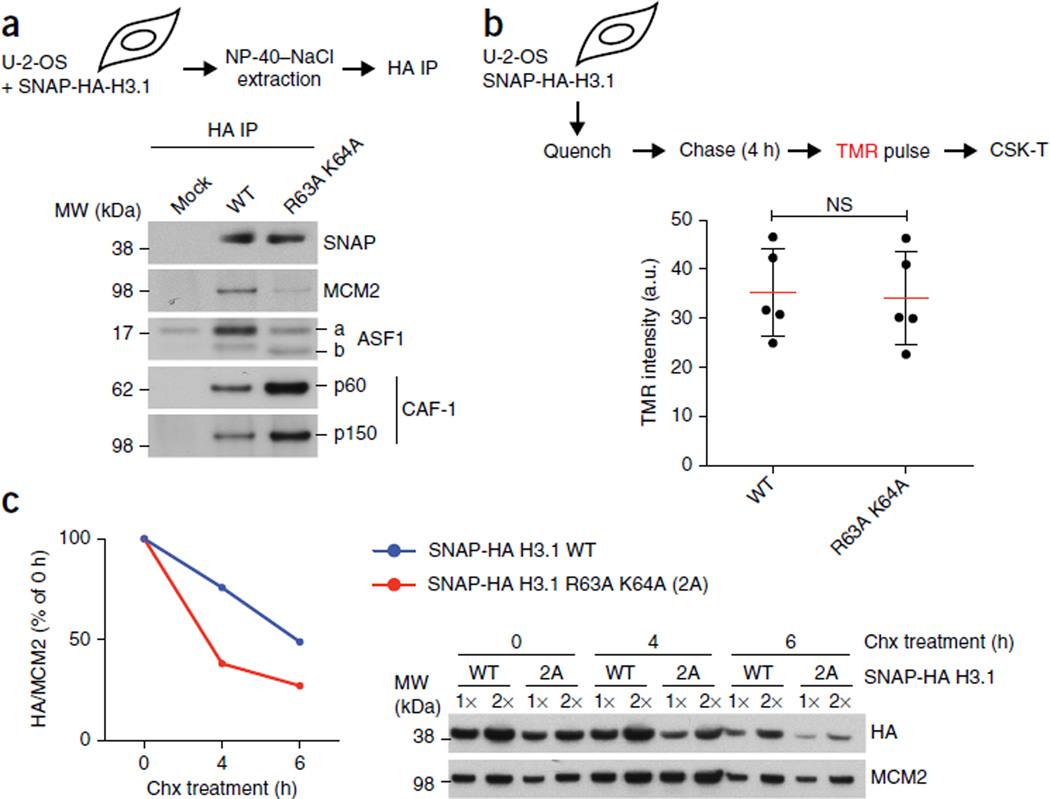
MCM2 binding stabilizes non-nucleosomal H3.1–H4. (a) IP of SNAP-HA-H3.1 WT or R63A K64A mutant from transfected cells (input material for IP in Supplementary Fig. 5a). (b) Replication-coupled H3.1 incorporation, measured by SNAP-tag fluorescent tetramethylrhodamine (TMR) labeling in stable cells expressing SNAP-HA-H3.1 WT or R63A K64A. Top, schematic showing quenching of old SNAP-HA-H3.1 to block labeling; incorporation of newly synthesized histones for 4 h before TMR labeling; and preextraction with CSK 0.5% Triton X-100 (CSK-T). Bottom, dot plot showing mean (red lines) TMR intensities are from five independent cell cultures and experiments, each including five technical replicates including more than 5,000 TMR-positive cells (representative micrographs in Supplementary Fig. 6a). Error bars (black lines), s.d.; NS, not significant by two-tailed unpaired t test; a.u., arbitrary units. (c) Stability of non-nucleosomal H3.1 WT and R63A K64A. Left, quantification, with SNAP-HA-H3.1 levels shown relative to MCM2. Right, western blot. Samples are soluble histones extracted from stable SNAP-HA-H3.1 WT or mutant cell lines synchronized in S phase and treated with hydroxyurea (HU) and cycloheximide (chx) to inhibit replication-coupled histone deposition and protein synthesis, respectively. One representative experiment out of three is shown. Uncropped images of gels are shown in Supplementary Data Set 1.