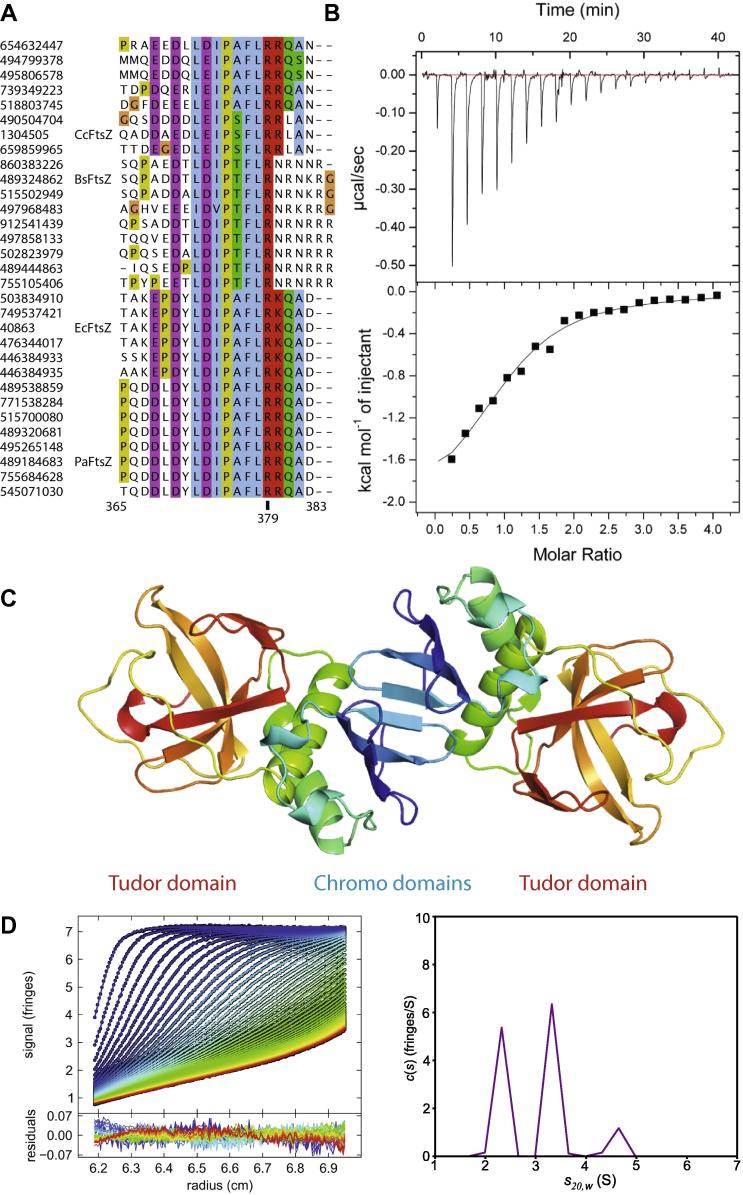
Fig. 2.
(A) Multiple sequence alignment (Clustal Omega, www.clustal.org) showing conservation within the C-terminal tails of various FtsZ proteins. A maximum of ten sequences were collected from each BLAST search with FtsZ sequences from E. coli, B. subtilis, C. crescentus and P. aeruginosa (Ec, Bs, Cc, PaFtsZ) sampled evenly up to the point where percentage of sequence cover and identify dropped below 95% and 40%, respectively. Totally conserved arginine residue 379 is highlighted; numbering corresponds to EcFtsZ. (B) Binding of FtsZ peptide to ZapC. Raw heats measured during injections of FtsZ peptide into ZapC by ITC (top) were integrated and fitted to a single-site binding model (bottom) yielding a stoichiometry of 1.0 with an enthalpy of −2.1 kcal/mol and KD of 32 ± 6 μM. (C) ZapC dimer as suggested by crystal packing and PISA (PDBe PISA; www.ebi.ac.uk/pdbe/pisa) [30]. The strands of the N-terminal chromo domain come together to form an eight-stranded sheet. (D) Left: analysis of the oligomeric state of ZapC from sedimentation velocity analytical ultracentrifugation. Interference scans (symbols) and best-fit c(s) model at different points in time indicated by colour temperature with residuals to the fit below. Right: c(s) sedimentation coefficient distribution showing peaks for monomer, dimer and higher oligomer.