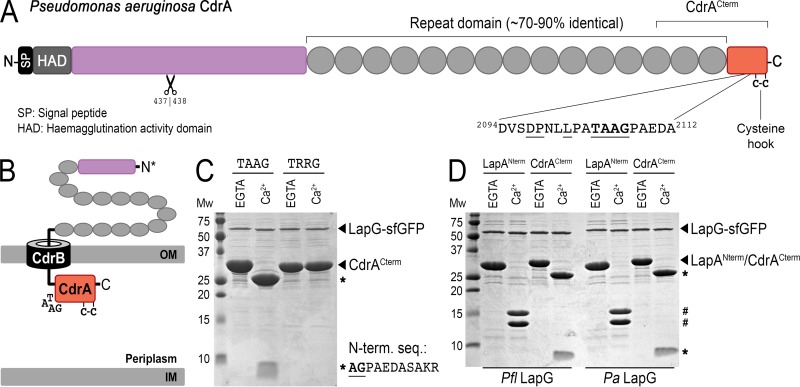
FIG 2.
LapG proteolysis of P. aeruginosa CdrACterm. (A) Primary structure and domain organization of CdrA, depicting the location of the LapG cleavage site and the putative C-terminal “cysteine hook.” A second, N-terminal proteolytic site identified previously to be between residues 437 and 438 (9) is also depicted. (B) Cartoon model for the localization of CdrA at the outer membrane via its cognate TPS transporter CdrB, in which the C terminus of CdrA resides in the periplasm while the N terminus is excreted into the extracellular space. Such a topological orientation is the opposite of that of LapA-like proteins, in which the C terminus is extracellular (Fig. 1A). (C) Proteolysis of purified CdrACterm by purified LapG-sfGFP produces 23-kDa and 5-kDa fragments (indicated by asterisks) in the presence of calcium but not EGTA. The sequence of the first 10 residues of the 5-kDa proteolytic fragment determined by Edman degradation is shown. A variant of CdrACterm in which the double-alanine motif is mutated to arginine residues (TRRG) prevents proteolysis. (D) Proteolytic processing of CdrACterm and LapANterm by P. fluorescens (left) and P. aeruginosa (right) LapG-sfGFP demonstrates functional conservation across the two systems. LapANterm and CdrACterm proteolytic products are indicated by # and *, respectively.