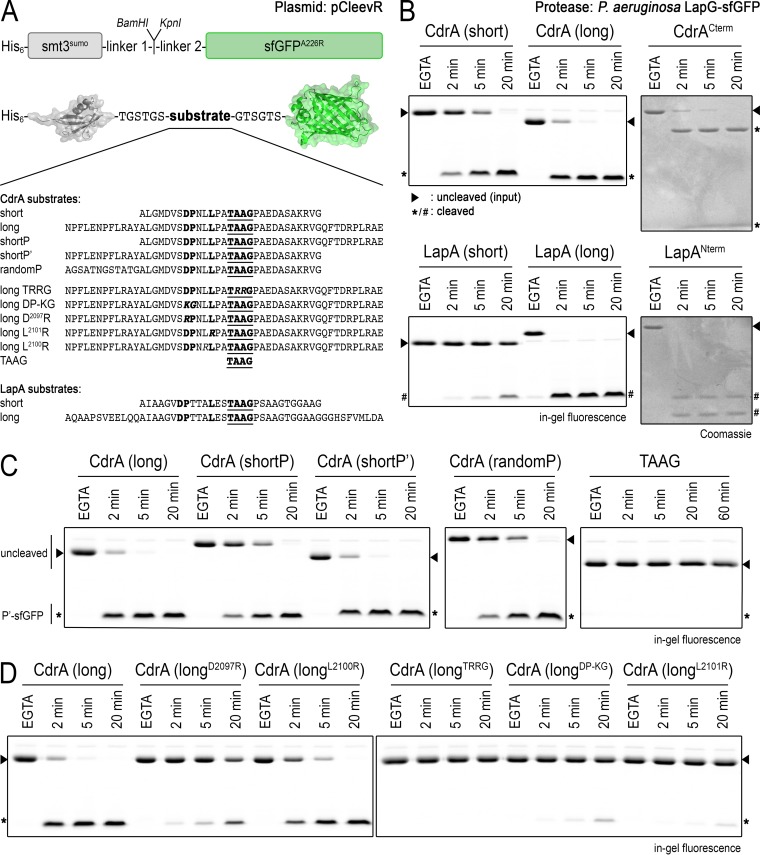
FIG 3.
Characterization of LapG specificity with the pCleevR assay. (A) Primary structure of pCleevR constructs in which various CdrA and LapA sequences containing their respective LapG cleavage sites are flanked N terminally by a His6-tagged SUMO protein and C terminally by sfGFP A226R. (B) Length of the pCleevR sequence influences rate of cleavage by LapG. CdrAshort/LapAshort (as defined in panel A) sequences are cleaved more slowly than CdrAlong/LapAlong sequences, which are processed by LapG at approximately the same rate as CdrACterm and LapANterm. Cleavage products are indicated by asterisks (for CdrA) and number symbols (for LapA). Intact substrates are indicated with triangles. (C) Assessment of the relative rates of cleavage of CdrAshortP′ and CdrAshortP substrates compared with the CdrAlong substrate indicates that residues N terminal to the cleavage site are responsible for this increased rate of proteolysis. The fact that CdrArandomP, which has the same length and number of residues flanking the cleavage site as CdrAshortP′ but is of unrelated sequence, is cleaved more slowly than CdrAshortP′ indicates that the enhanced rate is sequence dependent. The motif TAAG, in and of itself, is not sufficient for LapG targeting (right). Note that CdrAlong migrates with an aberrantly smaller size than CdrAshort in SDS-PAGE. (D) Single-amino-acid substitutions in CdrAlong identify residues D2097, P2098, and L2101 but not L2100 as crucial for LapG-mediated cleavage. As with the TRRG variant of CdrACterm (Fig. 2C), the analogous TRRG CdrAlong variant is not cleaved by LapG.