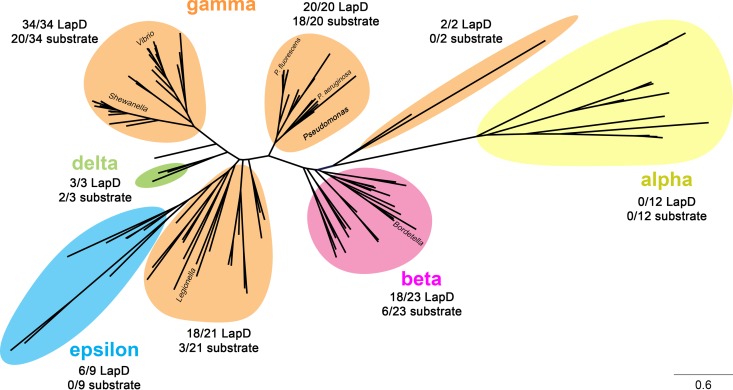
FIG 8.
Phylogenetic tree of LapG orthologs, displaying the diversity of the LapDG signaling system in bacteria. For this analysis, 5,220 bacterial genomes were downloaded from the NCBI database and initially searched for a putative LapG-like protease (identified by the conserved DX9WX12DCED[YF]X3KX20–40HX12–17LD motif). These genomes were subsequently searched for a LapD-like protein (motif F[DE]X[GS]X[YF]X20–40PXW[FLIVM]X16–18GWX47–51[RP]L) and a LapG-like substrate possessing a DPX2–3LX2[TA]AAG motif. LapG sequences were aligned by Clustal Omega (25) and analyzed phylogenetically by maximum likelihood with the JTT model for amino acid substitutions (26). Colored shadings highlight the class of bacteria to which individual species belong. Beside each shaded group, the corresponding number of species containing a LapD ortholog and a putative LapG substrate is indicated relative to the number of LapG orthologs within this group. Note that for many species, while LapD and LapG orthologs can be identified, no readily apparent LapA- or CdrA-like proteolysis motif was found. This observation suggests that other novel LapG targets may be present in these microbes.