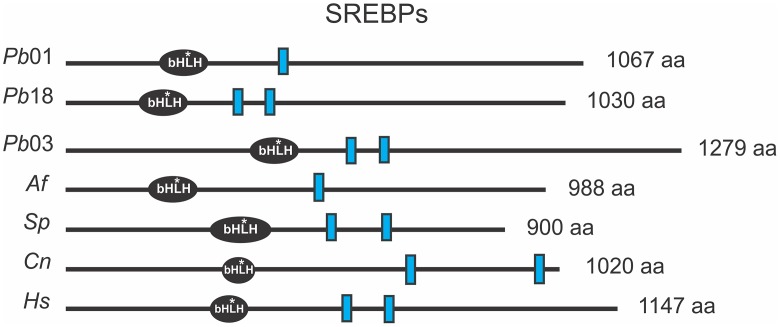
Fig 5. Predicted protein domains for SREBPs component orthologs.
The amino acid sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov/) to Paracoccidioides Pb01 (XP_002794199); Pb03 (KGY15961); Pb18 (XP_010758341); Aspergillus fumigatus (XP_749262); Schizosaccharomyces pombe (NP_595694); Cryptococcus neoformans (XP_567526) and Homo sapiens (P36956). The SREBPs bHLH (basic helix-loop- helix leucine zipper DNA-binding domain) protein domains and the length of each protein (indicated on the right of each protein in amino acids [aa]) were predicted by SMART tool (http://smart.embl-heidelberg.de) and the transmembrane segments (blue rectangles) were predicted using the Phobius (http://phobius.sbc.su.se/) and SACS MEMSAT2 Prediction softwares (http://www.sacs.ucsf.edu/cgi-bin/memsat.py). *: conserved tyrosine residue specific to the SREBP family of bHLH transcription factors.