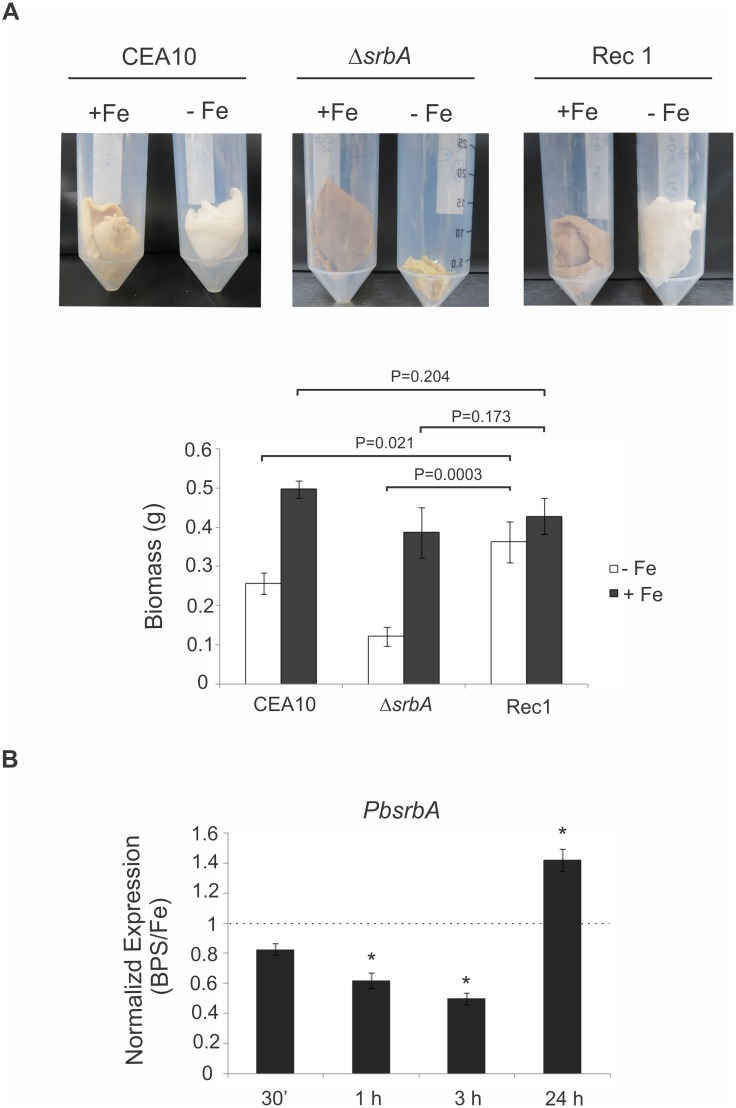
Fig 9. PbsrbA restores the deficient growth of the A. fumigatus srbA mutant under iron starvation.
(A) A total of 108 conidia of each wild type (CEA10), ΔsrbA and Rec1 (PbsrbA) strains of A. fumigatus in 50 ml liquid GMM was monitored after 48 h of growth at 37°C under iron starvation (-Fe) and iron availability (+Fe, 30 μM). The biomass production (dry mass) was compared among them, and expressed in grams (g). The data represent the mean ± standard deviation of biological triplicates. The difference between reconstituted strain and mutant/ wild-type strains was statistically significant during -Fe but not +Fe (t-test; P ≤ 0.05). (B) The analysis of transcripts abundance of PbsrbA was performed using qRT-PCR when yeast cells of Paracoccidioides Pb01 were submitted to iron sufficiency (30 μM) and deprivation (iron chelator bathophenanthrolinedisulfonate, BPS [50 μM]) culture media. Total RNAs were extracted and treated with DNAse. The cDNAs molecules were synthesized and qRT-PCR performed. The values that were plotted on the bar graph were normalized against the expression data that were obtained from the no iron addition condition (fold change). The data are expressed as the mean ± standard deviation of the triplicates. *statistically significant data as determined by Student’s t-test (P ≤ 0.01).