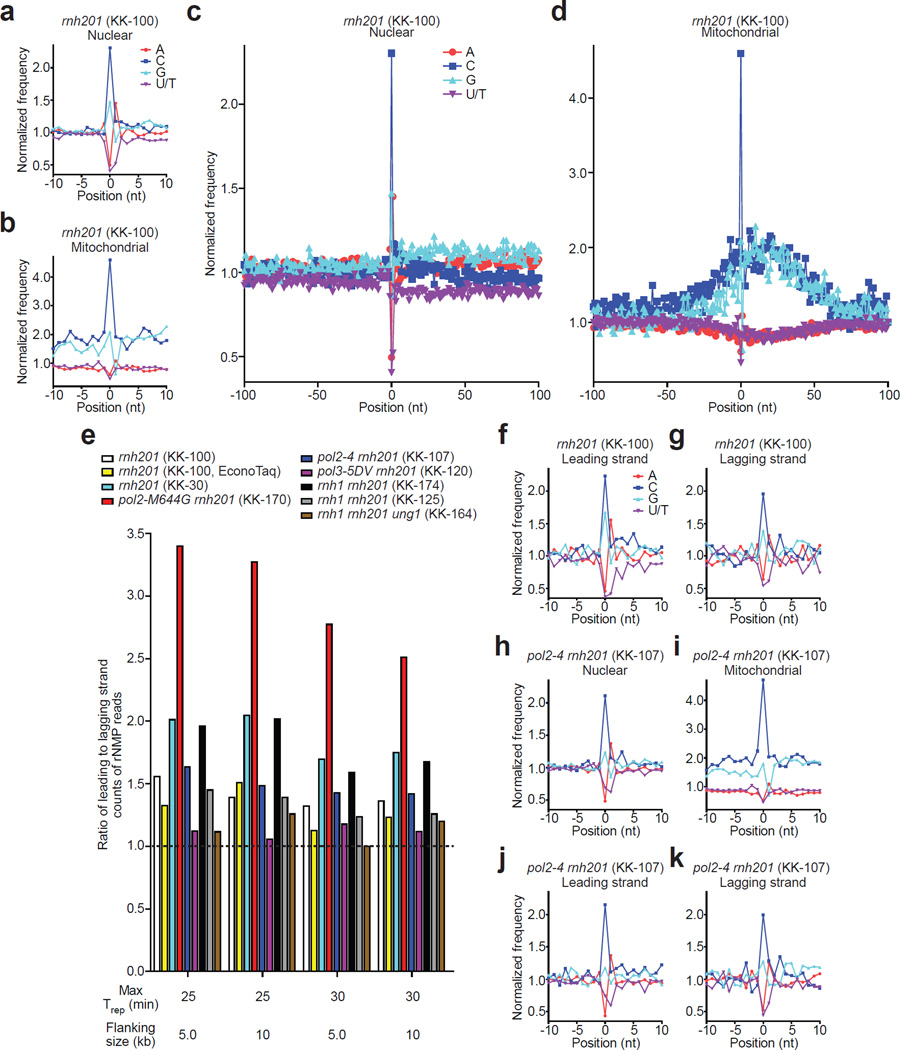
Figure 3. Identity and sequence contexts of rNMP incorporation in S. cerevisiae genome.
(a,b) Normalized nucleotide frequencies relative to mapped positions of sequences from Ribose-seq library of rnh201Δ (KK-100) cells. Position 0 is the rNMP. (c, d), Zoom-out of frequencies. (e) Ratios of rNMPs on newly-synthesized leading to lagging strand for all Ribose-seq libraries. Early firing ARSs selected by their replication timing (Trep) were investigated for two different flanking sizes. (f,g) Normalized nucleotide frequencies relative to mapped sequence positions in newly-synthesized leading and lagging strands from the rnh201Δ (KK-100) library. ARSs with Trep of no longer than 25 min were selected with flanking size of 10 kb. (h–k) Normalized nucleotide frequencies relative to mapped sequence positions from a pol24rnh201Δ (KK-107) library. Reads were mapped to (a,c,h) nuclear genome, (b,d,i) mitochondrial genome, (f,j) leading strand or (g,k) lagging strand.