Abstract
The microbial nitrogen cycle is one of the most complex and environmentally important element cycles on Earth and has long been thought to be mediated exclusively by prokaryotic microbes. Rather recently, it was discovered that certain eukaryotic microbes are able to store nitrate intracellularly and use it for dissimilatory nitrate reduction in the absence of oxygen. The paradigm shift that this entailed is ecologically significant because the eukaryotes in question comprise global players like diatoms, foraminifers, and fungi. This review article provides an unprecedented overview of nitrate storage and dissimilatory nitrate reduction by diverse marine eukaryotes placed into an eco-physiological context. The advantage of intracellular nitrate storage for anaerobic energy conservation in oxygen-depleted habitats is explained and the life style enabled by this metabolic trait is described. A first compilation of intracellular nitrate inventories in various marine sediments is presented, indicating that intracellular nitrate pools vastly exceed porewater nitrate pools. The relative contribution by foraminifers to total sedimentary denitrification is estimated for different marine settings, suggesting that eukaryotes may rival prokaryotes in terms of dissimilatory nitrate reduction. Finally, this review article sketches some evolutionary perspectives of eukaryotic nitrate metabolism and identifies open questions that need to be addressed in future investigations.
Keywords: nitrogen-cycle, nitrate respiration, denitrification, DNRA, diatoms, foraminifers, gromiids, fungi
Introduction
Nitrate is one of the major nutrients for microbial and plant life on planet Earth. It is the most oxidized form of fixed N-compounds, abundant in many aquatic habitats, and of high importance for both assimilatory and dissimilatory nitrogen metabolism.
Nitrate can also be stored inside living cells at concentrations by far exceeding ambient concentrations, a trait that is known for several prokaryotic and eukaryotic phyla (e.g., Dortch et al., 1984; Fossing et al., 1995; McHatton et al., 1996; Schulz et al., 1999; Lomas and Glibert, 2000; Needoba and Harrison, 2004; Risgaard-Petersen et al., 2006; Mußmann et al., 2007; Piña-Ochoa et al., 2010a; Kamp et al., 2011; Bernhard et al., 2012a; Coppens et al., 2014; Stief et al., 2014).
Apparently, the ability to store nitrate intracellularly is widely distributed within the eukaryotic tree of life (Figure 1). Extra- and intracellular nitrate serves for both assimilation and dissimilation. In assimilatory nitrate reduction, ammonium is produced and subsequently incorporated into biomass to build up e.g., proteins and nucleic acids. Dissimilatory nitrate reduction is a process for energy conservation, in which nitrate is used as an electron acceptor in the (near) absence of oxygen (e.g., Fewson and Nicholas, 1961; Strohm et al., 2007; Kraft et al., 2011; Thamdrup, 2012). Dissimilatory nitrate reduction and nitrate storage in particular are physiological life traits that provide microbes with environmental flexibility (i.e., metabolic activity under both oxic and anoxic conditions) and resource independence (i.e., anaerobic metabolism without immediate nitrate supply), respectively. Such life traits are especially important in environments that are temporarily anoxic and/or nitrate-free and they may have developed as a “life strategy” in both prokaryotes and eukaryotes (Figure 2).
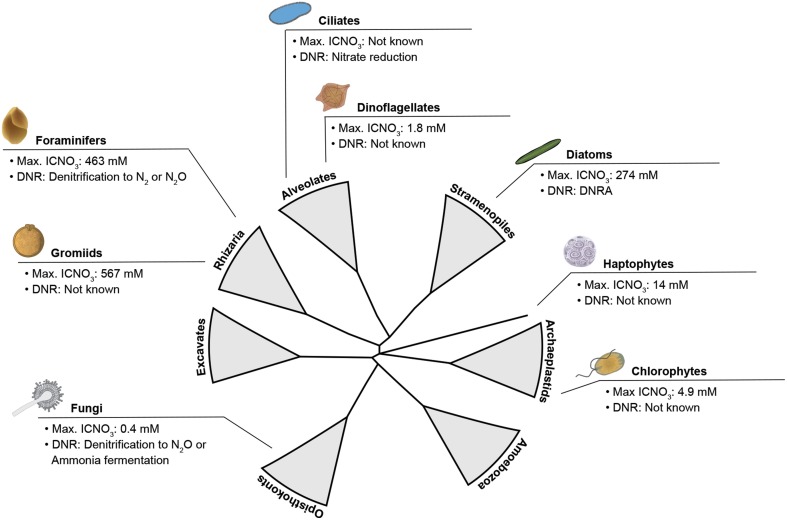
Figure 1.
Schematized eukaryotic tree of life emphasizing the wide distribution of lineages known to store nitrate intracellularly and use it for dissimilation. Maximum intracellular nitrate concentrations (max. ICNO3) and pathways of dissimilatory nitrate reduction (DNR) are given for taxonomic groups tested positive for either trait. DNRA, Dissimilatory Nitrate Reduction to Ammonium. DNR data compiled from Finlay et al. (1983) (ciliates), Shoun and Tanimoto (1991) (fungi), Zhou et al. (2002) (fungi), Risgaard-Petersen et al. (2006) (foraminifers), Kamp et al. (2011) (diatoms). Max. ICNO3 data compiled from Dortch et al. (1984) (dinoflagellates, haptophytes), Lomas and Glibert (2000) (chlorophytes), Piña-Ochoa et al. (2010a) (gromiids), Piña-Ochoa et al. (2010b) (foraminifers), Kamp et al. (2011) (diatoms), Stief et al. (2014) (fungi). Tree topology adapted from Worden et al. (2015).
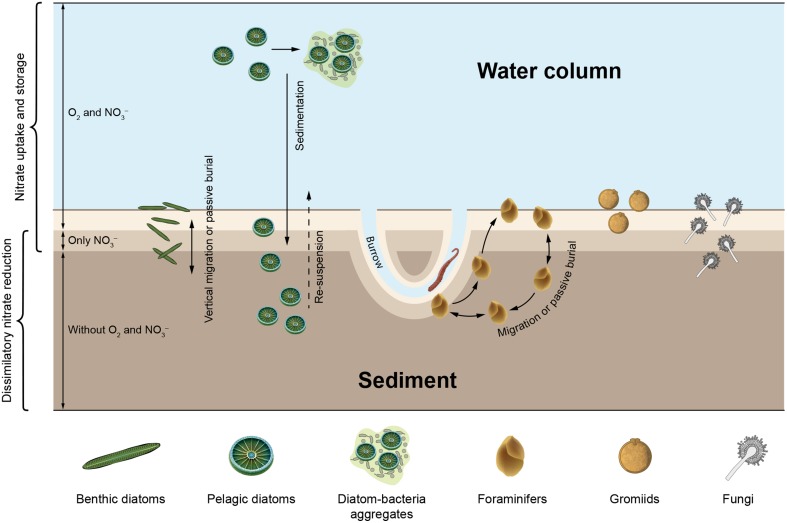
Figure 2.
Marine microbial eukaryotes known to take up ambient nitrate for intracellular storage and/or dissimilatory nitrate reduction drawn in a conceptual scheme of a natural environment with oxic and anoxic compartments. Organisms and environmental compartments are stylized and not to scale. Scenarios: (1) Benthic diatoms and foraminifers migrate actively up and down between oxic and anoxic sediment layers, or are buried in deep, anoxic sediment layers by e.g., macrofaunal activities, (2) Foraminifers move through different sediment layers and might re-fill their nitrate stores at “hotspots” of nitrate in deeper sediment layers, e.g., macrofaunal burrows, (3) Gromiids reside at the sediment surface or in anoxic subsurface layers, (4) Fungi grow in various sediment layers, (5) Pelagic diatoms sink onto the sediment after phytoplankton blooms and are re-suspended due to spring storms or macrofaunal activities, and (6) Pelagic diatoms are exposed to hypoxic or anoxic conditions inside sinking diatom-bacteria aggregates. Aside from the spatial separation into oxic and anoxic compartments, temporal variation of oxygen availability in the bottom water or inside macrofaunal burrows causes sudden shifts from oxic to anoxic conditions (and back) that may influence nitrate uptake and dissimilatory nitrate reduction by microbial eukaryotes (not shown).
Dissimilatory nitrate reduction pathways such as nitrate reduction to nitrite, denitrification, or Dissimilatory Nitrate Reduction to Ammonium (DNRA; Box 1), are well-studied in prokaryotes (i.e., Bacteria and Archaea). Prokaryotes are an integral part of the microbial nitrogen cycle; and due not least to the increasing use of fertilizers and the subsequent pollution of rivers, estuaries, and coastal waters, they have been the focus of many research activities for nearly a 100 years (e.g., Kluyver and Donker, 1926; Fewson and Nicholas, 1961; Zumft, 1997). In contrast, research on eukaryotes that can switch from oxygen to (intracellular) nitrate for anaerobic energy metabolism when (temporarily) exposed to anoxia or hypoxia is still in its infancy. However, the list of eukaryotes so far found to reduce nitrate dissimilatorily includes major global players such as benthic and pelagic marine diatoms, foraminifers, and fungi (Figure 1), which suggests a quantitative impact on nitrogen cycling at least in the marine realm. The first eukaryote that has been found to respire with nitrate in the absence of oxygen was, however, the freshwater ciliate Loxodes sp., which survives anoxic conditions in lakes through nitrate reduction to nitrite (Finlay et al., 1983). Later on, soil fungi were shown to be capable of incomplete denitrification to nitrous oxide and nitrate reduction to ammonium (Box 1; Shoun and Tanimoto, 1991; Takaya et al., 1999). Further, the fungus Aspergillus terreus isolated from a seasonal oxygen minimum zone (OMZ) in the Arabian Sea was shown to perform DNRA under anoxic conditions (Stief et al., 2014). In 2006, it was demonstrated that certain benthic foraminiferal species perform intracellular accumulation of nitrate, which is subsequently denitrified (Risgaard-Petersen et al., 2006). Only recently, it was discovered that the most important phototrophic group of microbial eukaryotes, the diatoms, also possess a dissimilatory nitrate metabolism. Both a benthic and a pelagic diatom have been shown to perform DNRA after sudden shifts to darkness and anoxia (Kamp et al., 2011, 2013).
Box 1. Pathways of dissimilatory nitrate reduction in eukaryotes.
Denitrification: Reduction of nitrate via nitrite, nitric oxide, and nitrous oxide to dinitrogen ( NO → N2O → N2) with organic or inorganic electron donors. Complete denitrification serves as an efficient N-removal pathway in the environment. Incomplete denitrification may begin and end at various points in the reaction sequence and, for example, produce the greenhouse gas N2O.
Dissimilatory Nitrate Reduction to Ammonium (DNRA): Reduction of nitrate via nitrite to ammonium ( ) with organic or inorganic electron donors. DNRA does not contribute to N-removal, but rather recycles fixed N in the environment.
Ammonia Fermentation: Reduction of nitrate to ammonium coupled to the oxidation of organic electron donors to acetate and substrate-level phosphorylation. Ammonia fermentation has the same effect on N-cycling in the environment as DNRA.
This review summarizes the current knowledge of (i) the ecophysiology of marine microbial eukaryotes that store nitrate intracellularly and thus are potentially involved in dissimilatory nitrate reduction and (ii) the environmental impact of nitrate storage and dissimilatory nitrate reduction by marine microbial eukaryotes on the nitrogen cycle of our oceans. The review further evaluates some evolutionary perspectives, and closes with a more general discussion of open questions that might inspire further research.
Ecophysiology
Nitrate storage
Diatoms
Among the marine eukaryotic microbes discussed here, intracellular nitrate storage was first described for diatoms, in which intracellular concentrations can by far exceed ambient concentrations (i.e., nitrate in the surrounding seawater). The range of intracellular nitrate concentrations varies according to species and environmental conditions. Many benthic and pelagic diatoms accumulate nitrate intracellularly in concentrations up to a few 100 mM, but concentrations can be as low as zero to a few mM (Dortch et al., 1984; Lomas and Glibert, 2000; Needoba and Harrison, 2004; Høgslund, 2008; Kamp et al., 2011, 2013; Coppens et al., 2014).
Nitrate uptake by diatoms has been shown to be temperature-dependent (decreasing with increasing temperature), is so far documented to occur in oxic conditions only, and uptake rates vary between species (e.g., Raimbault and Mingazzini, 1987; Lomas and Glibert, 1999, 2000; Villareal et al., 1999; Tantanasarit et al., 2013). Lomas and Glibert (2000) measured nitrate uptake rates of 18–310 fmol cell−1 h−1 for six different species grown at moderate ambient nitrate concentrations (< 40 μM). This rate can be even higher at very high ambient nitrate concentrations (Tantanasarit et al., 2013).
Intracellular nitrate is generally thought to be located in vacuoles. In the plant vacuole of Arabidopsis thaliana, for example, nitrate is accumulated via an /H+ exchanger (Martinoia et al., 1981; De Angeli et al., 2006). Evidence for a similar mechanism in unicellular eukaryotes is missing so far.
Nitrate storage in diatoms has long been assumed to serve assimilation exclusively (e.g., Dortch et al., 1984; Lomas and Glibert, 2000), probably because diatoms are mostly found in oxic habitats where nitrate is not needed as an alternative electron acceptor for dissimilation. The first hint for dissimilatory nitrate reduction in diatoms was a correlation between the nitrate storage capacity and the survival time of benthic and pelagic diatoms after sudden shifts to dark and anoxic conditions (Kamp et al., 2011). However, intracellular nitrate is used up within hours and is not replenished from ambient nitrate under anoxic conditions (see below). Lomas and Glibert (1999) also hypothesized that some diatom populations take up nitrate in excess of nutrient requirements because its reduction may serve as a sink for electrons during transient periods of imbalance between light energy harvesting and utilization.
Foraminifers
Foraminifers may store nitrate at concentrations >15,000 times the environmental nitrate concentrations (Risgaard-Petersen et al., 2006), probably in vacuoles (Bernhard et al., 2012a), and the measured intracellular nitrate pool varies among nitrate-storing foraminifers from ca. 0.1 mM to >375 mM (Piña-Ochoa et al., 2010a). This variation seems to reflect different physiological and environmental conditions rather than phylogenetic constraints because considerable intraspecific variation is observed among the species that are considered to be nitrate collectors (Piña-Ochoa et al., 2010a; Koho et al., 2011; Bernhard et al., 2012b). The ability to store nitrate at concentrations above environmental concentrations is so far found within the orders Allogromiida, Miliolida, Rotaliida, and Textulariida (Piña-Ochoa et al., 2010a; Bernhard et al., 2012a), and seems to be a common trait for foraminifers from very diverse benthic marine environments, such as OMZs, hypoxic basins, continental slopes, shelf sediments, and coastal sediments (Piña-Ochoa et al., 2010a; Bernhard et al., 2012a). Interestingly, the trait is not restricted to species which often occur in anoxic microhabitats, such as e.g., Globobulimina turgida, but is also found in species living in oxic habitats (e.g., Cassidulina carinata and Pyrgo elongata), and in opportunistic species (e.g., Bolivina subaenariensis and Uvigerina mediterranea; Piña-Ochoa et al., 2010a). 15N labeling experiments performed on Ammonia beccarii, Bolivina argentea, Buliminella tenuata, G. turgida*, Fursenkoina cornuta and Nonionella stella have shown that nitrate is taken up directly from the environment (Risgaard-Petersen et al., 2006; Koho et al., 2011; Bernhard et al., 2012b; Nomaki et al., 2014), and is not produced internally. The mechanism for nitrate uptake as well as the storage mode is at present unknown, but it must involve an active transport system, as nitrate is moved across the cell membrane against a large concentration gradient. From a thermodynamic point of view, the process of nitrate uptake is exergonic and therefore requires an investment of energy by the organism. G. turgida*, for instance, may accumulate nitrate internally to well above 10 mM in environments where the environmental concentration is less than 20 μM (Risgaard-Petersen et al., 2006). The Gibbs free energy (ΔG) for nitrate transport across the cell membrane at these conditions is >+15 kJ mol−1 according to equations in Harold (1986). It is evident therefore that nitrate accumulation among foraminifers can only be a sustainable strategy, if required to sustain processes that are essential for the survival of the organism. It has been shown that the nitrate-respiring foraminifer G. turgida can survive for up to 56 days of anoxia from respiration of its internal nitrate pool (Piña-Ochoa et al., 2010b), and the building and maintenance of an intracellular nitrate pool might be seen as an insurance that enables the organisms to sustain an active metabolism even when suitable external electron acceptors are absent in the environment (see below).
Gromiids
Like the foraminifers, gromiids belong to the Rhizaria (Burki et al., 2010; Sierra et al., 2013), and their intracellular nitrate concentrations can also reach >100 mM, which exceeds the ambient nitrate concentration by several orders of magnitude (Piña-Ochoa et al., 2010a). The ability to accumulate nitrate at these high concentrations appears to be ubiquitous for the gromiids, as it has been found for individuals sampled from hard-bottom substrates, shelf sediments in temperate and arctic regions, as well as in the OMZ along the coastline of Peru (Piña-Ochoa et al., 2010a). The physiology of the gromiids has only been superficially studied and neither the mechanism behind nitrate accumulation, nor its link to any metabolic pathway has been investigated so far. It is possible that the gromiid-nitrate association represents a system that is functionally different from that of benthic foraminifers because gromiids generally are described as surface dwellers, and thus not buried in anoxic sediment layers like many foraminifers (Jepps, 1926; Hedley and Bertaud, 1962; Arnold, 1972; Matz et al., 2008; da Silva and Gooday, 2009; Rothe et al., 2011).
Fungi
To date, only a single strain of A. terreus isolated from a marine sediment has been shown to store nitrate intracellularly and use it for dissimilatory nitrate reduction mainly to ammonium (Stief et al., 2014). The intracellular nitrate concentration in this strain reached up to 0.4 mM. Unfortunately, intracellular nitrate storage has not been studied in the large number of soil fungi and yeasts capable of dissimilatory nitrate reduction (Takaya et al., 1999; Maeda et al., 2015). Fungi in general, however, do possess cellular vacuoles and nitrate transporters (Klionsky et al., 1990; Navarro et al., 2006) and are able to take up nitrate from the environment at high rates and store it in vacuoles (e.g., 9 nmol mg−1 dry weight min−1 in Aspergillus nidulans; Unkles et al., 2004).
Ciliates
For the only ciliate known to perform dissimilatory nitrate reduction, Loxodes sp. (Finlay et al., 1983), intracellular nitrate storage has not been reported.
Chlorophytes, dinoflagellates, and haptophytes
Marine phytoplankton belonging to these eukaryotic lineages has mainly been investigated with respect to uptake and assimilation of nitrate and ammonium, but intracellular nitrate storage is also reported occasionally (e.g., Dortch et al., 1984; Lomas and Glibert, 2000). The chlorophyte Dunaliella tertiolecta stored nitrate at 2.7–4.9 mM in one study (Lomas and Glibert, 2000), but had intracellular nitrate concentrations below the detection limit in another study (Dortch et al., 1984). Similarly, the dinoflagellate Amphidinium carterae stored 0–1.8 mM nitrate (Dortch et al., 1984), while the dinoflagellate Prorocentrum minimum did not store nitrate (Lomas and Glibert, 2000). Among the haptophytes, Isochrysis galbana stored 0.3–13.9 mM nitrate (Dortch et al., 1984) and Pavlova lutheri only stored 0.1–0.2 mM nitrate (Lomas and Glibert, 2000). Clearly, more investigations focusing on intracellular nitrate storage in these lineages are needed.
Dissimilatory nitrate reduction
Diatoms
So far, the benthic diatom Amphora coffeaeformis and the pelagic diatom Thalassiosira weissflogii have been shown to reduce nitrate dissimilatorily. Both diatom strains perform the pathway Dissimilatory Nitrate Reduction to Ammonium (DNRA), as demonstrated with 15N labeling experiments in axenic strains (Kamp et al., 2011, 2013). The DNRA rates of these two diatoms are in the range of 2–3 fmol N cell−1 h−1 during the first hours after exposure to dark and anoxic conditions. However, DNRA rates become significantly lower after only a few hours, which mirrors the rapid consumption of intracellular nitrate after shifts to darkness and anoxia. Thus, diatoms probably use the intracellular nitrate, and its dissimilatory reduction via DNRA, either for short-term survival or for entering a resting stage.
To date, genes involved in dissimilatory nitrate reduction have not been identified in diatoms, but only in denitrifying soil fungi (see below). Intriguingly, fungi use enzymes that are usually involved in assimilatory nitrate reduction in a dissimilatory mode (Takasaki et al., 2004). This could also be true for diatoms. Assimilatory nitrate reductases, nitrate transporters, and components of a nitrate-sensing system have only recently been identified in diatom genomes (Armbrust et al., 2004; Bowler et al., 2008). Identification of functional genes involved in dissimilatory nitrate reduction in diatoms would provide genetic evidence for this metabolic pathway in diatoms.
Foraminifers
Direct measurements of nitrate reduction activity associated with nitrate-storing foraminifers have demonstrated a capacity for complete denitrification of to N2 (Risgaard-Petersen et al., 2006; Høgslund, 2008; Piña-Ochoa et al., 2010a; Bernhard et al., 2012b). Some species (e.g., Bolivina plicata, Bolivina seminuda, Valvulineria cf. laevigata, Stainforthia sp.), however, seem to lack nitrous oxide reductase and reduce nitrate only to nitrous oxide (Piña-Ochoa et al., 2010a).
At present, denitrification rates for only 11 different species within the Rotaliida order have been determined. The observation of elevated δ15NNO3 and δ18ONO3 values in the intracellular nitrate pool within allogromiid foraminifers from the Santa Barbara Basin (Bernhard et al., 2012a) has demonstrated nitrate reduction capacity associated with members of the Allogromiida order, yet rate measurements in this order are still missing. Rates estimated for the Rotaliida with N2O-microsensors (Risgaard-Petersen et al., 2006; Høgslund et al., 2008; Piña-Ochoa et al., 2010a,b) or 15 amendments (Risgaard-Petersen et al., 2006; Bernhard et al., 2012b) fall in the range of 1.7–83 pmol N cell−1 h−1, and great intraspecific variation is observed. There is a tendency for a log-log relationship between the denitrification rate and biovolume of the organisms, so that large organisms have higher rates than smaller ones, as seen also for foraminiferan cell-specific oxygen respiration rates (Geslin et al., 2011), but the current database is too limited for strong conclusions to be drawn. In general, individual denitrification rates are much lower than the corresponding oxygen respiration rates (Piña-Ochoa et al., 2010a) and it has been suggested that denitrification is an auxiliary metabolism used for cell maintenance, food collection, and locomotion during temporary stays in oxygen-free environments, whereas oxygen might be required for growth and reproduction (Piña-Ochoa et al., 2010a,b).
The genes behind the foraminiferan denitrification pathway have not been elucidated. It has, however, been shown with microscopy (Risgaard-Petersen et al., 2006) and experiments applying bacteria-specific antibiotics to denitrifying foraminifers (Bernhard et al., 2012b) that for some species (e.g., B. argentea and G. turgida*) the foraminifers themselves, and not only the associated prokaryotes, are performing the denitrification reaction. Denitrification in a nitrate-storing allogromiid foraminifer from the Santa Barbara Basin, however, appears to be performed by prokaryotic endobionts and not the eukaryote, as demonstrated by sequence analyses and GeneFISH (Bernhard et al., 2012a). Given the widespread distribution of nitrate-accumulating and denitrifying foraminifers within diverse phylogenetic orders, specific investigations of each group are needed, at best on the genomic level, to confirm or reject the presence of eukaryotic denitrification. It is obvious from the Santa Barbara study that a capacity for nitrate accumulation is not necessarily coupled to a capacity of the eukaryote to utilize this directly for energy conservation through e.g., denitrification.
Fungi
The best-studied fungi species capable of dissimilatory nitrate reduction are the two soil-living plant pathogens Fusarium oxysporum and Cylindrocarpon tonkinense (Shoun and Tanimoto, 1991; Usuda et al., 1995). The majority of terrestrial fungi, some ectomycorrhizal fungi, and many of the yeast strains screened since the initial discovery of “fungal denitrification” also tested positive for this trait (Tsuruta et al., 1998; Prendergast-Miller et al., 2011; Mothapo et al., 2013; Maeda et al., 2015). A key feature of “fungal denitrification” is the absence of the last reduction step of the denitrification pathway, which makes fungi very important nitrous oxide (N2O) producers in soils (Laughlin and Stevens, 2002; Crenshaw et al., 2008; Chen et al., 2014). Fungi isolated from aquatic ecosystems have received much less attention in terms of dissimilatory nitrate reduction, and conclusive experiments with 15 labeling have been made for only one single strain of A. terreus isolated from sediment in the seasonal oxygen minimum zone of the Arabian Sea (Stief et al., 2014). This strain also has a high N2O yield (approximately 15% of the total amount of N produced), but the main product of its nitrate reduction activity is ammonium (up to 83%, equivalent to 175 nmol N g−1 protein h−1), which is also the case for a number of soil fungi (Zhou et al., 2002). The underlying metabolic pathway has been termed “ammonia fermentation” a process which couples the oxidation of ethanol to acetate, and the reduction of nitrate to ammonium, to substrate-level phosphorylation (Box 1; Takaya, 2009). Thus, two pathways of dissimilatory nitrate reduction have evolved in fungi, apparently also within individual species (e.g., F. oxysporum; Zhou et al., 2002). The prevalence of either pathway is controlled by ambient oxygen levels, with hypoxic and anoxic levels triggering “fungal denitrification” and “ammonia fermentation,” respectively (Takaya, 2009).
Key genes of “fungal denitrification” have been identified and sequenced (Kizawa et al., 1991; Kim et al., 2009). Nitrite reduction to nitric oxide (NO) is mediated by a copper-containing nitrite reductase (NirK), while the reduction step from NO to N2O is mediated by the cytochrome P450 nitric oxide reductase (P450nor). Nitrous oxide reductases are generally absent in fungi, which explains why N2O instead of N2 is the final product of “fungal denitrification” (Takaya, 2009). Dissimilatory nitrate reductases can be present in some denitrifying fungi species, but are less well-characterized than NirK and p450nor (Takaya, 2009). Hence, the minimal denitrification pathway in fungi only comprises the two-step reduction of nitrite to N2O. The stepwise reduction of nitrate to ammonium in fungal “ammonia fermentation” is apparently mediated by assimilatory nitrate (NiaD) and nitrite reductases (NiiA) used in a dissimilatory context, i.e., it is coupled to the fermentation of ethanol to acetate (Takasaki et al., 2004). Meanwhile, several primer sets for the fungal NirK and p450nor have been developed and used for screening fungal isolates for their capability of dissimilatory nitrate reduction (Kim et al., 2010; Maeda et al., 2015; Mothapo et al., 2015; Wei et al., 2015). The availability of these primer sets will enable the detection of denitrifying fungi in environmental samples without prior isolation, cultivation, and functional testing.
Ciliates
The freshwater ciliate Loxodes sp. survives anoxic conditions in lakes through dissimilatory nitrate reduction to nitrite (Finlay et al., 1983; Aleya et al., 1992). A link was made between the anaerobic metabolism of the ciliate and a higher number of mitochondria per cell and a greater surface area of cristae inside the mitochondria compared to specimens exposed to oxic conditions (Finlay et al., 1983; Finlay, 1985). To date, the gene encoding the nitrate reductase has not been identified.
Chlorophytes, dinoflagellates, and haptophytes
None of the aforementioned nitrate-storing representatives of these eukaryotic lineages (see Nitrate Storage) has been tested for dissimilatory nitrate reduction under anoxic conditions so far.
Habitats and life style
Dissimilatory nitrate reduction by prokaryotes and eukaryotes typically occurs in environments in which the availability of oxygen and nitrate is variable in space and time. Stable environments are spatially structured into zones with/without oxygen and/or nitrate availability, while dynamic environments are temporally structured into phases with/without oxygen and/or nitrate availability. In aquatic ecosystems, such conditions can be found in sediments, around animal burrows in sediments, in the root zone of aquatic plants, in low-oxygen water bodies, and inside sinking organic aggregates.
In sediments with stable redox stratification, oxygen, as the most favorable electron acceptor in terms of energy, is consumed within the top few millimeters (Revsbech et al., 1980). Nitrate penetrates slightly deeper into the sediment where it is used as an alternative electron acceptor when oxygen is depleted (Sweerts and de Beer, 1989). Microbes that are able to store nitrate intracellularly may thrive well below the nitrate penetration depth, but need to fill up their nitrate stores occasionally. Large sulfur bacteria couple vertical migration behavior to uptake and storage of nitrate at the sediment surface and dissimilatory use of intracellular nitrate deeper in the sediment (e.g., Fossing et al., 1995). Benthic foraminifers and diatoms, both of which are capable of migrating inside sediments, can be abundant well below the nitrate penetration depth (Figure 2; Risgaard-Petersen et al., 2006; Stief et al., 2013). Benthic diatoms exhibit a vertical migration rhythm that is coupled to diurnal and tidal cycles (Consalvey et al., 2004), while foraminifers migrate more erratically or directed to oxygen gradients (Alve and Bernhard, 1995; Geslin et al., 2004; Koho et al., 2011). As a consequence of their migration behavior, benthic diatoms and foraminifers are exposed to elevated ambient nitrate and oxygen levels whenever they reach the sediment surface. Deeper in the sediment where they find shelter from predation and erosion (Kingston, 1999), diatoms and foraminifers face the absence of ambient nitrate and oxygen.
Oxygen and nitrate concentration gradients in sediments can experience rapid and pronounced changes caused by disturbance events. Short-term oxygen and nitrate pulses occur in animal burrows that reach deep into anoxic sediment layers and are intermittently irrigated with oxygen- and nitrate-rich surface water (Kristensen et al., 1991; Wenzhöfer and Glud, 2004). During the resting phase of the animals, oxygen is depleted faster than nitrate, which allows for short-term dissimilatory nitrate reduction in the immediate surrounding of the burrow (Stief and de Beer, 2006). The root zone of aquatic plants exhibits oxygen and nitrate dynamics that are similar to animal burrows, albeit due to periodic changes in the photosynthetic activity of the plant (Frederiksen and Glud, 2006). In the light, roots release oxygen into the surrounding sediment and stimulate nitrate production by microbial nitrification, while in the dark, nitrate is depleted due to dissimilatory nitrate reduction activities (Risgaard-Petersen and Jensen, 1997). It has been suggested that the nitrate-storing and sulfide-oxidizing Thioploca ingrica are able to exploit nitrate pulses in animal burrows to fill up their nitrate stores (Høgslund et al., 2010). Benthic diatoms and foraminifers are often abundant in this dynamic microenvironment in which conditions conducive to nitrate uptake and dissimilatory nitrate reduction alternate (Alve and Bernhard, 1995; Steward et al., 1996).
Hypoxic or anoxic water bodies in which nitrate is available may also host dissimilatory nitrate reduction mediated by eukaryotes. The ciliate Loxodes sp. is abundant just below the oxic-anoxic interface of stratified lakes, where it reduces nitrate dissimilatorily to nitrite (Finlay et al., 1983; Aleya et al., 1992). Marine pelagic diatoms can move up and down through the water column by controlling their buoyancy (Armbrust, 2009) and are thereby exposed to varying ambient nitrate and oxygen levels (Villareal et al., 1993). Rapid and large-scale transport of diatoms through the water column of the oceans occurs when diatoms and bacteria aggregate to form “marine snow” (Thornton, 2002). Sinking organic aggregates also exhibit internal gradients of oxygen concentration due to microbial respiration and transport limitation of oxygen (Ploug et al., 1997). Under dark conditions (i.e., at night or when aggregates sink out of the photic zone) the center of aggregates may become anoxic, which allows for dissimilatory nitrate reduction (Klawonn et al., 2015). Pelagic diatoms finally sink onto the seafloor, where they still host a large inventory of intracellular nitrate (Lomstein et al., 1990) and may survive in dark, anoxic sediment layers for decades (Härnström et al., 2011), thus far longer than the intracellular nitrate pool would last.
Environmental impact
Inventory of intracellular nitrate pools
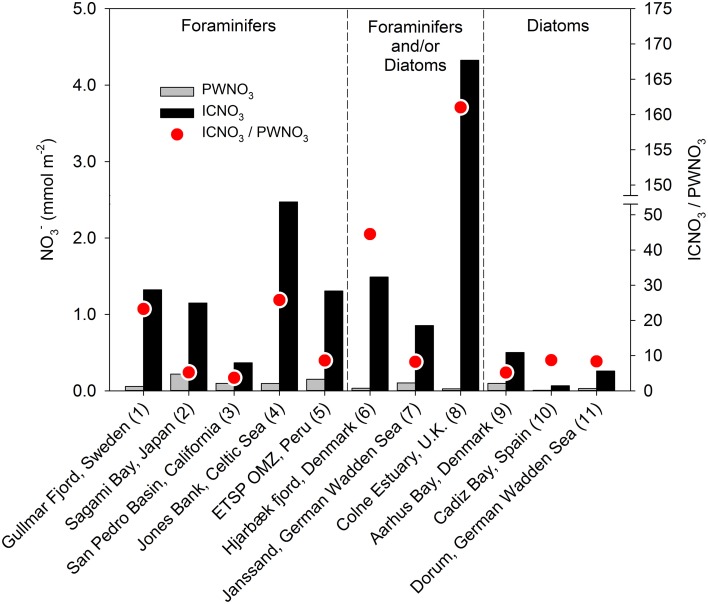
The presence of nitrate-storing eukaryotes in sediments leads to large inventories of nitrate that vastly exceed the porewater nitrate contents in some environments, equivalent to what can be observed in sediments colonized with the sulfide-oxidizing bacteria Thioploca sp. or Beggiatoa sp. (Jørgensen and Gallardo, 1999; Sayama, 2001). These intracellular nitrate pools (ICNO3 pools) are measured with a diverse set of methods, such as freeze-thaw cycling, boiling, whole-core squeezing, and centrifugation of environmental samples, which all aim at lysing nitrate-storing cells (e.g., Lomstein et al., 1990; Risgaard-Petersen et al., 2006; Prokopenko et al., 2011; Larsen et al., 2013). In sediments from the Gullmar Fjord, Sweden, for instance, nitrate dissolved in the sediment porewater (PWNO3) accounted for less than 4% of the total nitrate pool (Risgaard-Petersen et al., 2006). The remaining nitrate, as extracted by boiling the sediment, was most likely present in eukaryotic cells since neither Beggiatoa, nor Thioploca was present. The sediment was inhabited by the nitrate-storing foraminifer Globobulimina pseudospinescens, and the cell-bound nitrate was significantly correlated with the abundance of this organism. However, the intracellular nitrate pool of G. pseudospinescens only accounted for approximately 20% of total nitrate in the sediment, leaving open the possibility that other nitrate-storing foraminifers or diatoms, gromiids, and fungi were present. Settled phytoplankton with nitrate-storing representatives among the diatoms, chlorophytes, dinoflagellates, and haptophytes may contribute to the sedimentary ICNO3 pool that is not accounted for by benthic eukaryotes and prokaryotes. Meanwhile, large sedimentary ICNO3 pools were ascribed to the presence of benthic foraminifers in various marine ecosystems (Figures 3A, 4; Table S1). Intracellular nitrate pools were ~4–26 times larger than porewater nitrate pools (Figure 4; Glud et al., 2009; Prokopenko et al., 2011; Glock et al., 2013; Larsen et al., 2013). In three additional studies, it was assumed that both foraminifers and/or diatoms contribute to the ICNO3 pool in marine sediments (Figures 3A,B, 4; Table S1; Høgslund et al., 2010; Marchant et al., 2014; Papaspyrou et al., 2014).
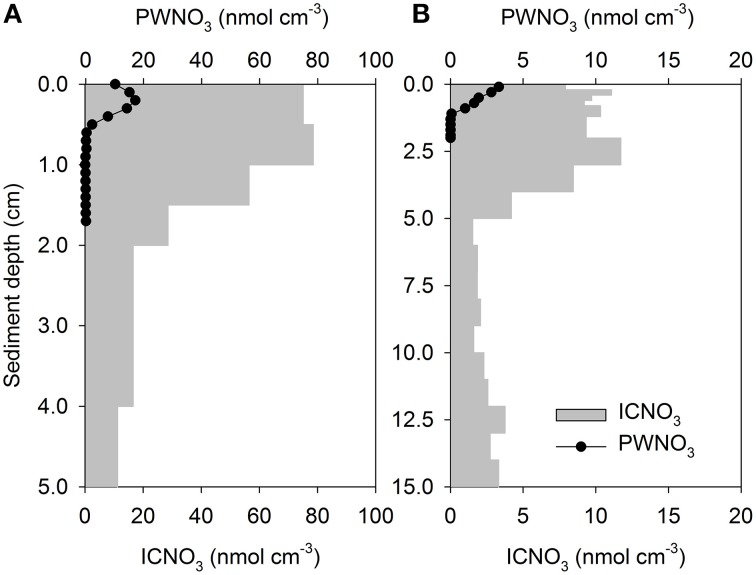
Figure 3.
Vertical profiles of porewater nitrate (PWNO3) and intracellular nitrate (ICNO3) in (A) foraminifer-inhabited and (B) diatom-inhabited marine sediments. Nitrate concentrations are expressed per cm3 of sediment. Note different scales. Data compiled from (A) Risgaard-Petersen et al. (2006) and (B) Heisterkamp et al. (2012).
Figure 4.
Inventories of porewater nitrate (PWNO3) and intracellular nitrate (ICNO3) in various marine sediments. Only studies in which the sedimentary ICNO3 pool is ascribed to nitrate-storing foraminifers and/or diatoms are considered. Data compiled from (1) Risgaard-Petersen et al. (2006), (2) Glud et al. (2009), (3) Prokopenko et al. (2011), (4) Larsen et al. (2013), (5) Glock et al. (2013), (6) Høgslund et al. (2010), (7) Marchant et al. (2014), (8) Papaspyrou et al. (2014), (9) Lomstein et al. (1990), (10) García-Robledo et al. (2010), and (11) Heisterkamp et al. (2012). Details on data extraction can be found in Table S1.
In a number of coastal sediments, the total ICNO3 pool has been exclusively assigned to microalgae, in particular to pelagic diatoms that have settled onto the sediment surface (Lomstein et al., 1990) and to benthic diatoms that reside in intertidal sediments (García-Robledo et al., 2010; Heisterkamp et al., 2012; Stief et al., 2013). In natural settings, diatom-associated ICNO3 is diagnosed as a congruent distribution of ICNO3 and fucoxanthin, the marker pigment of diatoms (Stief et al., 2013). In diatom-dominated sediments, the ratio of ICNO3-to-PWNO3 tends to be lower (~5–9) than in foraminifer-dominated sediments (Figure 4; Table S1), but more data need to be collected to confirm this preliminary observation. The pronounced seasonality in intertidal communities of the temperate zone also entails seasonal changes of the diatom-associated ICNO3 pool with high and low values in the cold and warm season, respectively (Stief et al., 2013). It is currently not known whether other nitrate-storing eukaryotes show similar seasonal variation of their ICNO3 contents.
The in situ turnover of the sedimentary ICNO3 pool is indicated by elevated δ15NNO3 and δ18ONO3 values (Prokopenko et al., 2011; Bernhard et al., 2012b). These isotope ratios further increase when isolated foraminifers are incubated under anoxic conditions, which confirms dissimilatory nitrate reduction activity fueled by ICNO3 (Bernhard et al., 2012b). Estimated turnover times of ICNO3 vary between ~12 h and ≥1 month (Risgaard-Petersen et al., 2006; Høgslund, 2008; Glud et al., 2009; Bernhard et al., 2012b), which is considerably slower than the turnover of PWNO3 of only 2–4 h, which is determined in sediments inhabited by foraminifers (Glud et al., 2009; Larsen et al., 2013). This slow turnover of ICNO3 by foraminifers and other nitrate-storing eukaryotes has implications for rate measurements based on 15 incubations (Høgslund, 2008). The labeled and non-labeled nitrate pools may not readily mix within short incubation times, which leads to a significant underestimation of benthic denitrification rates determined with the isotope pairing technique (Nielsen, 1992). Additionally, the unintended release of ICNO3 from eukaryotic cells into the sediment porewater due to crude extraction techniques simulates concentration peaks that might be mistaken for nitrate production zones.
Estimates of eukaryotic dissimilatory nitrate reduction
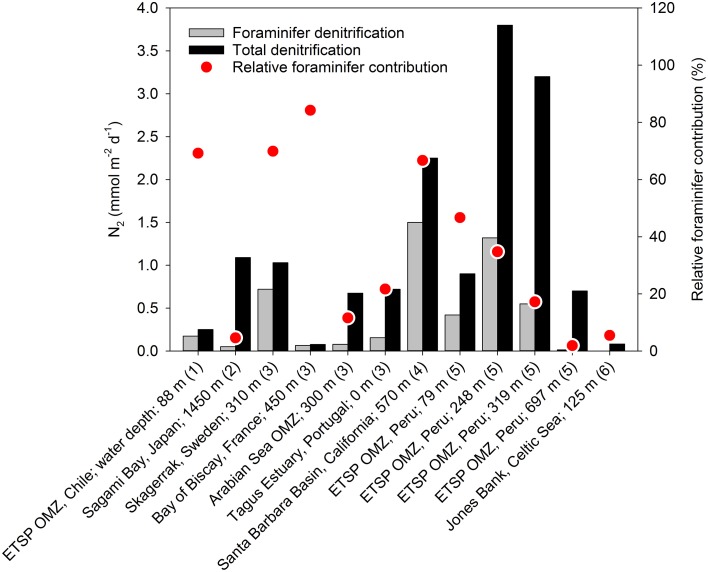
The quantitative role of eukaryote-associated nitrate reduction has only been addressed for foraminifers. The contribution of foraminiferal denitrification to the total loss of combined nitrogen from marine sediments has been estimated for various benthic settings (Høgslund, 2008; Glud et al., 2009; Piña-Ochoa et al., 2010a; Bernhard et al., 2012b; Glock et al., 2013). Apparently, foraminifers may contribute substantially to benthic denitrification and in some environments they even surpass the contribution from prokaryotes (Figure 5).
Figure 5.
Foraminiferan denitrification and total denitrification in various benthic marine environments. Data compiled from (1) Høgslund et al. (2008), (2) Glud et al. (2009), (3) Piña-Ochoa et al. (2010a), (4) Bernhard et al. (2012b), (5) Glock et al. (2013), and (6) Larsen et al. (2013).
Foraminiferan denitrification is in general estimated from the in situ abundance of live foraminifers and laboratory-based estimates of denitrification rates for individual species, whereas total denitrification is estimated from 15N-enrichment studies, from porewater profiles of nitrate, or from analyzing the distribution of δ15NNO3 in natural settings (Groffman et al., 2006). This approach involves a high degree of uncertainty as (i) there is only limited information about the diversity of foraminiferal denitrification activity (data from only 11 species are available), (ii) the cell-specific activity is typically measured at conditions far from natural environmental conditions, and (iii) foraminiferal denitrification is typically not included in standard techniques used for measuring total denitrification (see above). Therefore, present reports on foraminiferal contribution to benthic denitrification activity should be considered as preliminary attempts. There is certainly a need for methodologies that capture the in situ denitrification activity mediated by eukaryotes vs. prokaryotes.
Evolution
The last decade of research has shown that the eukaryotes have their evolutionary roots in a largely anoxic ocean (Anbar and Knoll, 2002; Martin et al., 2003). Pronounced atmospheric oxygenation occurred around 2.4 to 2.1 billion years ago as a consequence of oxygenic photosynthesis, and is named the Great Oxidation Event (GOE). For a few years, it has been debated whether the first production and local accumulation of oxygen might already have happened 2.7–3.2 billion years ago (Brocks et al., 1999; Lyons et al., 2014; Satkoski et al., 2015). Even after the GOE, however, it was not until ~600 million years ago that widespread oxygenation of the deep ocean occurred (Canfield, 1998; Canfield et al., 2008; Lyons et al., 2014). During this transition period of ~1.8 billion years, oxygen was first produced in oceanic microhabitats, which allowed nitrification, the critical aerobic pathway in the nitrogen cycle that produces nitrate, to proceed. Thus, nitrate was present in the oxic microniches of the Proterozoic ocean, and became available for dissimilatory nitrate reducers living on the edge of the nitrate production zones (Fennel et al., 2005; Canfield et al., 2010).
The occurrence of dissimilatory nitrate reduction in very distantly related eukaryotes (Figure 1) raises the question whether the genes involved were present in the single eukaryotic common ancestor that emerged in the largely anoxic Proterozoic ocean, or, alternatively, whether the dissimilatory nitrate reduction pathways we observe today exhibit multiple origins. Neither of the two hypotheses excludes the mitochondrion as the location for eukaryotic nitrate dissimilation. The mitochondrial proteome is a mosaic assemblage of proteins where some are traced to the last mitochondrial ancestor within the Alphaproteobacteria, and some have been acquired from other prokaryotes and eukaryotes through the course of evolution (Gray, 2015).
There is ample evidence that contemporary mitochondria-related organelles are derived from the same ancestral organelle (Mentel and Martin, 2008; van der Giezen, 2011) and the genes involved in dissimilatory nitrate reduction could have been introduced during this event. The suggested ancestor of the mitochondria within the Alphaproteobacteria (Yang et al., 1985; Williams et al., 2007; Gray, 2015) is a representative of a metabolically versatile class that contains facultative anaerobic species capable of running both aerobic respiration and denitrification, e.g., Paracoccus denitrificans, which is discussed as a candidate for the protomitochondrion (John and Whatley, 1975; Gray, 2015). The metabolic blueprint provided by the protomitochondrial Alphaproteobacteria has been extensively modified and today we observe diverse functions and biochemical pathways tied to mitochondria in different eukaryotic lineages (Müller et al., 2012). Investigations of mitochondrial proteomes in nitrate-reducing eukaryotes may show diverse routes of acquisition of the proteins involved in the pathways. This seems to be the case at least for the fungi.
The fungal nitrate respiration is tied to the mitochondrion and details of the denitrification pathway of the soil fungus F. oxysporum have largely been resolved. The conversion of nitrite to nitric oxide is coupled to the mitochondrial electron transport chain and ATP synthesis and involves a copper-containing nitrite reductase, NirK (Kobayashi et al., 1996; Kim et al., 2009; Long et al., 2015). Because the distribution of the eukaryotic NirK gene is systematic and follows the eukaryotic phylogeny, it is suggested that the trait of nitrite reduction evolved from a single ancestor and was carried into the eukaryotic domain in the endosymbiotic event leading to the evolution of the mitochondrion (Kim et al., 2009; Shoun et al., 2012). Apparently though, the fungal reduction of nitric oxide to nitrous oxide has a different origin (Shoun et al., 2012). This reduction step is mediated by a P450 nitric oxide reductase that is found in both the mitochondria and in the cytoplasm, and it seems that this part of the denitrification pathway has been acquired by lateral gene transfer (Kizawa et al., 1991).
Moving from the fungi in the Opisthokonta to the ciliates in the Alveolata, we also find that dissimilatory nitrate reduction by the ciliate Loxodes sp. is presumably affiliated with the mitochondria (Finlay et al., 1983; Finlay, 1985). Gene sequences of this nitrate reductase are, however, not available and it is, at the moment, not possible to draw conclusions about the origin of the trait.
Nitrate accumulation is dispersed throughout the foraminiferan phylogeny including allogromiid species, which likely evolved in the Neoproterozoic anoxic ocean and are considered to form a basal order within the foraminifers (Pawlowski et al., 2003). This might suggest that nitrate accumulation was present in the most recent common ancestor of foraminifers, albeit lost in some lineages. Usage of the intracellular nitrate may have evolved differently among foraminiferal lineages, since data at present show that the allogromiids share their nitrate with endobionts (Bernhard et al., 2012a), whereas some more recently evolved rotaliids invoke it in their own dissimilatory metabolic pathways (Risgaard-Petersen et al., 2006).
Genes for dissimilatory nitrate reduction in diatoms are not known, but diatoms have remarkable genomes with traces of multiple plastid endosymbiotic events allowing migration of genes from the plastid endobiont into the genome (Prihoda et al., 2012). A large number of genes also seem to be derived from bacteria by lateral gene transfer (Bowler et al., 2008; Armbrust, 2009), including genes for nitrite reductases that are targeted at the mitochondria (Allen et al., 2006). This knowledge of the chimeric diatom genomes paves the way for speculations on lateral transfers of genes involved in the dissimilatory use of nitrate, but the examination of such hypotheses requires further sequencing and bioinformatics efforts.
The extensive lack of information on the genes and enzymes driving dissimilatory nitrate reduction among the eukaryotes also means that we cannot close in on the time of its evolutionary origin. It can be established, however, that an anaerobic energy metabolism coupled to nitrate reduction was possible in the environmental settings at the time of origin of ascomycete fungi (Lücking et al., 2009; Prieto and Wedin, 2013) and foraminifers (Pawlowski et al., 2003; Groussin et al., 2011). Diatoms evolved only 250 million years ago (Sims et al., 2006; Armbrust, 2009) and existing hypotheses of diatom origins tend to agree that the pre-diatom or “Ur-diatom” developed in shallow marine (and thus more oxygenated) environments (Sims et al., 2006; Medlin, 2011). It will be interesting to follow the evolutionary path of dissimilatory nitrate reduction among eukaryotes when more sequence data becomes available.
Open questions and further directions
The study of nitrate storage and dissimilatory nitrate reduction by eukaryotic microbes is still in its infancy. Finding nitrate reduction to nitrite by Loxodes sp. and denitrification by fungi were early milestones reached a few decades ago. More recently though, this research area has gained momentum by the discovery that diatoms and foraminifers are also capable of dissimilatory nitrate reduction coupled to intracellular nitrate storage. Future research activities should address open questions regarding the (i) phylogenetic diversity, (ii) physiology and genetics, and (iii) in situ importance of eukaryotic nitrate storage and dissimilatory nitrate reduction.
The known occurrence of nitrate storage and reduction in distant eukaryotic lineages (Figure 1) suggests that these physiological traits are even more widespread among eukaryotes than previously thought. In particular, those lineages for which nitrate storage has already been documented might be “hot candidates” for performing dissimilatory nitrate reduction under anoxic conditions, e.g., gromiids and dinoflagellates (Dortch et al., 1984; Piña-Ochoa et al., 2010a). There is a growing interest in microbial eukaryotes adapted to life under low-oxygen conditions (e.g., Stoeck et al., 2009; Edgcomb et al., 2011; Müller et al., 2012; Bernhard et al., 2014; Parris et al., 2014). An obvious research strategy is, thus, to test known and novel eukaryotic lineages from low-oxygen environments for their ability to store nitrate intracellularly (Step 1) and use nitrate as an alternative electron acceptor (Step 2).
Some fundamentals of the physiology, biochemistry, and genetics of eukaryotic nitrate storage and reduction are still unknown. While 15N labeling experiments have proven intracellular nitrate as an alternative electron acceptor in dissimilatory processes in fungi, foraminifers, and diatoms, there is still a role for intracellular nitrate in assimilation. The exact partitioning of intracellular nitrate between dissimilation and assimilation remains to be investigated in the diverse nitrate-storing and nitrate-reducing eukaryotes. Further unknowns concern the mechanism and energy requirements of nitrate uptake, the intracellular compartment of nitrate storage, and the spectrum of electron donors used for dissimilatory nitrate reduction (e.g., organic vs. inorganic, external vs. storage compounds).
The identification of genes that encode for enzymes involved in dissimilatory nitrate reduction by diatoms and foraminifers is a challenging task that needs particular attention. Knowledge of these genes will not only provide insights into the evolution and biochemistry of dissimilatory nitrate reduction in eukaryotes, but will also enable the development of molecular tools for cultivation-independent investigations directly in the environment. Lessons might be learned from the investigation of denitrifying fungi (Kim et al., 2009; Wei et al., 2015). Several diatom genomes have recently been sequenced, annotated, and interpreted in the context of the cellular nitrogen metabolism (Armbrust et al., 2004; Allen et al., 2006; Bowler et al., 2008). Furthermore, transcriptome sequencing projects of microbial eukaryotes are forthcoming and will provide a rich source of sequence information that can be screened for genes involved in eukaryotic dissimilatory nitrate reduction (Keeling et al., 2014).
The question of dissimilatory nitrate reduction mediated by putative bacterial symbionts in foraminifers (Bernhard et al., 2012a) may also be resolved as soon as eukaryotic and prokaryotic genes for this process can be distinguished. Diatoms and fungi are known to host bacterial endosymbionts too (Foster and Zehr, 2006; Kobayashi and Crouch, 2009), but so far there are no reports on an involvement of these symbionts in dissimilatory nitrate reduction. The relationship between endosymbiotic nitrate reducers and a nitrate-storing eukaryotic host is still enigmatic. It seems paradoxical that a host organism should spend energy to accumulate nitrate intracellularly and then leave it to a bacterial partner without any benefit to itself. In some foraminifers, however, endosymbiotic bacteria are known to use intracellular nitrate for synthesizing amino acids from which the eukaryotic host may benefit (Nomaki et al., 2014, 2015).
The quantitative role of eukaryotic dissimilatory nitrate reduction in the environment is highly uncertain. It has actually been calculated that denitrification by benthic foraminifers equals denitrification by prokaryotes in some marine sediments (Piña-Ochoa et al., 2010a), and calculations are yet to be done for the other eukaryotes. However, the differential measurement of eukaryotic and prokaryotic rates directly in the environment has not yet been achieved due to methodological constraints, and thus novel techniques that capture the uptake and dissimilatory reduction of nitrate in mixed microbial communities in situ need to be developed. Additionally, there is no consensus method available for measuring ICNO3 pools directly in the environment. It should be evaluated whether freeze-thaw cycling, boiling, whole-core squeezing, and centrifugation are equally efficient in lysing nitrate-storing cells in environmental samples. So far, these techniques also lack selectivity in terms of which taxonomic group contributes how much to the total ICNO3 pool.
Given the ubiquitous distribution and high abundance of diatoms, fungi, and foraminifers in marine ecosystems, eukaryotic nitrate storage and dissimilatory nitrate reduction definitely have the potential to contribute significantly to the marine nitrogen cycle. The products of the different pathways of eukaryotic dissimilatory nitrate reduction range from a harmless gas (i.e., dinitrogen) to a strong greenhouse gas (i.e., nitrous oxide) that will escape into the atmosphere. Other products like ammonium or nitrite might be further used by prokaryotes with important roles in nitrogen cycling, such as nitrifiers, denitrifiers, and anammox bacteria. Disentangling the network of nitrogen transformations by eukaryotes and prokaryotes will provide a more comprehensive picture of the marine nitrogen cycle than is currently available.
Author contributions
This review article was conceived, written, and edited by all authors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financially supported by a grant from the German Research Foundation (Deutsche Forschungsgemeinschaft) awarded to AK (KA3187/2-1), a grant from the Aarhus University Research Foundation awarded to SH (AU Ideas), and the Danish National Research Foundation (NR-P; DNRF104). Tinna Christensen is acknowledged for help with Figures 1, 2, Emma Hammarlund is thanked for critical comments on the manuscript, and Karen ní Mheallaigh is thanked for proofreading. The manuscript was improved by the critical comments of the reviewers.
Footnotes
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01492
References
- Aleya L., Hartmann H. J., Devaux J. (1992). Evidence for the contribution of ciliates to denitrification in a eutrophic lake. Eur. J. Protistol. 28, 316–321. 10.1016/S0932-4739(11)80238-5 [DOI] [PubMed] [Google Scholar]
- Allen A. E., Vardi A., Bowler C. (2006). An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr. Opin. Plant Biol. 9, 264–273. 10.1016/j.pbi.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Alve E., Bernhard J. M. (1995). Vertical migratory response of benthic foraminifera to controlled oxygen concentrations in an experimental mesocosm. Mar. Ecol. Prog. Ser. 116, 137–151. 10.3354/meps116137 [DOI] [Google Scholar]
- Anbar A. D., Knoll A. H. (2002). Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142. 10.1126/science.1069651 [DOI] [PubMed] [Google Scholar]
- Armbrust E. V. (2009). The life of diatoms in the world's oceans. Nature 459, 185–192. 10.1038/nature08057 [DOI] [PubMed] [Google Scholar]
- Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. 10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- Arnold Z. M. (1972). Observations on the biology of the protozoan Gromia oviformis. Univ. Calif. Publ. Zool. 100, 1–168. [Google Scholar]
- Bernhard J. M., Casciotti K. L., McIlvin M. R., Beaudoin D. J., Visscher P. T., Edgcomb V. P. (2012b). Potential importance of physiologically diverse benthic foraminifera in sedimentary nitrate storage and respiration. J. Geophys. Res. 117, G03002 10.1029/2012JG001949 [DOI] [Google Scholar]
- Bernhard J. M., Edgcomb V. P., Casciotti K. L., McIlvin M. R., Beaudoin D. J. (2012a). Denitrification likely catalyzed by endobionts in an allogromiid foraminifer. ISME J 6, 951–960. 10.1038/ismej.2011.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard J. M., Kormas K., Pachiadaki M. G., Rocke E., Beaudoin D. J., Morrison C., et al. (2014). Benthic protists and fungi of Mediterranean deep hypsersaline anoxic basin redoxcline sediments. Front. Microbiol. 5:605. 10.3389/fmicb.2014.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- Brocks J. J., Logan G. A., Buick R., Summons R. E. (1999). Archean molecular fossils and the early rise of eukaryotes. Science 285, 1033–1036. 10.1126/science.285.5430.1033 [DOI] [PubMed] [Google Scholar]
- Burki F., Kudryavtsev A., Matz M. V., Aglyamova G. V., Bulman S., Fiers M., et al. (2010). Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol. 10:377. 10.1186/1471-2148-10-377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield D. E. (1998). A new model for Proterozoic ocean chemistry. Nature 396, 450–453. 10.1038/24839 [DOI] [Google Scholar]
- Canfield D. E., Glazer A. N., Falkowski P. G. (2010). The evolution and future of earth's nitrogen cycle. Science 330, 192–196. 10.1126/science.1186120 [DOI] [PubMed] [Google Scholar]
- Canfield D. E., Poulton S. W., Knoll A. H., Narbonne G. M., Ross G., Goldberg T., et al. (2008). Ferruginous conditions dominated later neoproterozoic deep-water chemistry. Science 321, 949–952. 10.1126/science.1154499 [DOI] [PubMed] [Google Scholar]
- Chen H. H., Mothapo N. V., Shi W. (2014). The significant contribution of fungi to soil N2O production across diverse ecosystems. Appl. Soil Ecol. 73, 70–77. 10.1016/j.apsoil.2013.08.011 [DOI] [Google Scholar]
- Consalvey M., Paterson D. M., Underwood G. J. C. (2004). The ups and downs of life in a benthic biofilm: migration of benthic diatoms. Diatom Res. 19, 181–202. 10.1080/0269249X.2004.9705870 [DOI] [Google Scholar]
- Coppens J., Decostere B., Van Hulle S., Nopens I., Vlaeminck S. E., De Gelder L., et al. (2014). Kinetic exploration of nitrate-accumulating microalgae for nutrient recovery. Appl. Microbiol. Biotechnol. 98, 8377–8387. 10.1007/s00253-014-5854-9 [DOI] [PubMed] [Google Scholar]
- Crenshaw C. L., Lauber C., Sinsabaugh R. L., Stavely L. K. (2008). Fungal control of nitrous oxide production in semiarid grassland. Biogeochemistry 87, 17–27. 10.1007/s10533-007-9165-4 [DOI] [Google Scholar]
- da Silva A. A., Gooday A. J. (2009). Large organic-walled Protista (Gromia) in the Arabian Sea: density, diversity, distribution and ecology. Deep Sea Res. II 56, 422–433. 10.1016/j.dsr2.2008.12.027 [DOI] [Google Scholar]
- De Angeli A., Monachello D., Ephritikhine G., Frachisse J. M., Thomine S., Gambale F., et al. (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442, 939–942. 10.1038/nature05013 [DOI] [PubMed] [Google Scholar]
- Dortch Q., Clayton J. R., Jr., Thoresen S. S., Ahmed S. I. (1984). Species differences in accumulation of nitrogen pools in phytoplankton. Mar. Biol. 81, 237–250. 10.1007/BF00393218 [DOI] [Google Scholar]
- Edgcomb V., Orsi W., Taylor G. T., Vdacny P., Taylor C., Suarez P., et al. (2011). Accessing marine protists from the anoxic Cariaco Basin. ISME J. 5, 1237–1241. 10.1038/ismej.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennel K., Follows M., Falkowski P. G. (2005). The co-evolution of the nitrogen, carbon and oxygen cycles in the Proterozoic ocean. Amer. J. Sci. 305, 526–545. 10.2475/ajs.305.6-8.526 [DOI] [Google Scholar]
- Fewson C. A., Nicholas D. J. D. (1961). Utilization of nitrate by micro-organisms. Nature 190, 2–7. 10.1038/190002a0 [DOI] [PubMed] [Google Scholar]
- Finlay B. J. (1985). Nitrate respiration by Protozoa (Loxodes spp.) in the hypolimnetic nitrite maximum of a productive freshwater pond. Freshw. Biol. 15, 333–346. 10.1111/j.1365-2427.1985.tb00205.x [DOI] [Google Scholar]
- Finlay B. J., Span A. S. W., Harman J. M. P. (1983). Nitrate respiration in primitive eukaryotes. Nature 303, 333–336. 10.1038/303333a0 [DOI] [Google Scholar]
- Fossing H., Gallardo V. A., Jørgensen B. B., Hüttel M., Nielsen L. P., Schulz H., et al. (1995). Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature 374, 713–715. 10.1038/374713a0 [DOI] [Google Scholar]
- Foster R. A., Zehr J. P. (2006). Characterization of diatom-cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences. Environ. Microbiol. 8, 1913–1925. 10.1111/j.1462-2920.2006.01068.x [DOI] [PubMed] [Google Scholar]
- Frederiksen M. S., Glud R. N. (2006). Oxygen dynamics in the rhizosphere of Zostera marina: a two-dimensional planar optode study. Limnol. Oceanogr. 51, 1072–1083. 10.4319/lo.2006.51.2.1072 [DOI] [Google Scholar]
- García-Robledo E., Corzo A., Papaspyrou S., Jiménez-Arias J. L., Villahermosa D. (2010). Freeze-lysable inorganic nutrients in intertidal sediments: dependence on microphytobenthos abundance. Mar. Ecol. Prog. Ser. 403, 155–163. 10.3354/meps08470 [DOI] [Google Scholar]
- Geslin E., Heinz P., Jorissen F., Hemleben C. (2004). Migratory responses of deep-sea benthic foraminifera to variable oxygen conditions: laboratory investigations. Mar. Micropaleontol. 53, 227–243. 10.1016/j.marmicro.2004.05.010 [DOI] [Google Scholar]
- Geslin E., Risgaard-Petersen N., Lombard F., Metzger E., Langlet D., Jorissen F. (2011). Oxygen respiration rates of benthic foraminifera as measured with oxygen microsensors. J. Exp. Mar. Biol. Ecol. 396, 108–114. 10.1016/j.jembe.2010.10.011 [DOI] [Google Scholar]
- Glock N., Schönfeld J., Eisenhauer A., Hensen C., Mallon J., Sommer S. (2013). The role of benthic foraminifera in the benthic nitrogen cycle of the Peruvian oxygen minimum zone. Biogeoscience 10, 4767–4783. 10.5194/bg-10-4767-2013 [DOI] [Google Scholar]
- Glud R. N., Thamdrup B., Stahl H., Wenzhoefer F., Glud A., Nomaki H., et al. (2009). Nitrogen cycling in a deep ocean margin sediment (Sagami Bay, Japan). Limnol. Oceanogr. 54, 723–734. 10.4319/lo.2009.54.3.0723 [DOI] [Google Scholar]
- Gray M. W. (2015). Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 10133–10138. 10.1073/pnas.1421379112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groffman P. M., Altabet M. A., Böhlke J. K., Butterbach-Bahl K., David M. B., Firestone M. K., et al. (2006). Methods for measuring denitrification: diverse approaches to a difficult problem. Ecol. Appl. 16, 2091–2122. 10.1890/1051-0761(2006)016[2091:MFMDDA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Groussin M., Pawlowski J., Yang Z. (2011). Bayesian relaxed clock estimation of divergence times in foraminifera. Mol. Phyl. Evol. 61, 157–166. 10.1016/j.ympev.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Härnström K., Ellegaard M., Andersen T. J., Godhe A. (2011). Hundred years of genetic structure in a sediment revived diatom population. Proc. Natl. Acad. Sci. U.S.A. 108, 4252–4257. 10.1073/pnas.1013528108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold F. M. (1986). The Vital Force: A Study of Bioenergetics. New York, NY: Freeman and Co. [Google Scholar]
- Hedley R. H., Bertaud W. S. (1962). Electron-microscopic observations of Gromia oviformis (Sarcodina). J. Protozool. 9, 79–87. 10.1111/j.1550-7408.1962.tb02585.x [DOI] [Google Scholar]
- Heisterkamp I. M., Kamp A., Schramm A. T., de Beer D., Stief P. (2012). Indirect control of the intracellular nitrate pool of intertidal sediment by the polychaete Hediste diversicolor. Mar. Ecol. Prog. Ser. 445, 181–192. 10.3354/meps09464 [DOI] [Google Scholar]
- Høgslund S. (2008). Nitrate Storage as an Adaption to Benthic Life. Ph.D. thesis, University of Aarhus, Aarhus. [Google Scholar]
- Høgslund S., Nielsen J. L., Nielsen L. P. (2010). Distribution, ecology and molecular identification of Thioploca from Danish brackish water sediments. FEMS Microbiol. Ecol. 73, 110–120. 10.1111/j.1574-6941.2010.00878.x [DOI] [PubMed] [Google Scholar]
- Høgslund S., Revsbech N. P., Cedhagen T., Nielsen L. P., Gallardo V. A. (2008). Denitrification, nitrate turnover, and aerobic respiration by benthic foraminiferans in the oxygen minimum zone off Chile. J. Exp. Mar. Biol. Ecol. 359, 85–91. 10.1016/j.jembe.2008.02.015 [DOI] [Google Scholar]
- Jepps M. W. (1926). Contribution to the study of Gromia oviforinis Dujardin. Q. J. Microsc. Sci. 70, 701–719. [Google Scholar]
- John P., Whatley F. R. (1975). Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature 254, 495–498. 10.1038/254495a0 [DOI] [PubMed] [Google Scholar]
- Jørgensen B. B., Gallardo V. A. (1999). Thioploca spp.: filamentous sulfur bacteria with nitrate vacuoles. FEMS Microbiol. Ecol. 28, 301–313. 10.1016/S0168-6496(98)00122-6 [DOI] [Google Scholar]
- Kamp A., de Beer D., Nitsch J. L., Lavik G., Stief P. (2011). Diatoms respire nitrate to survive dark and anoxic conditions. Proc. Natl. Acad. Sci. U.S.A. 108, 5649–5654. 10.1073/pnas.1015744108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp A., Stief P., Knappe J., de Beer D. (2013). Response of the ubiquitous pelagic diatom Thalassiosira weissflogii to darkness and anoxia. PLoS ONE 8:e82605. 10.1371/journal.pone.0082605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J., Burki F., Wilcox H. M., Allam B., Allen E. E., Amaral-Zettler L. A., et al. (2014). The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. 10.1371/journal.pbio.1001889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Fushinobu S., Zhou S., Wakagi T., Shoun H. (2009). Eukaryotic nirK genes encoding copper-containing nitrite reductase: originating from the protomitochondrion? Appl. Environ. Microbiol. 75, 2652–2658. 10.1128/AEM.02536-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Fushinobu S., Zhou S., Wakagi T., Shoun H. (2010). The possible involvement of copper-containing nitrite reductase (NirK) and flavohemoglobin in denitrification by the fungus Cylindrocarpon tonkinense. Biosci. Biotechnol. Biochem. 74, 1403–1407. 10.1271/bbb.100071 [DOI] [PubMed] [Google Scholar]
- Kingston M. B. (1999). Wave effects on the vertical migration of two benthic microalgae: Hantzschia virgata var. intermedia and Euglena proxima. Estuaries 22, 81–91. 10.2307/1352929 [DOI] [Google Scholar]
- Kizawa H., Tomura D., Oda M., Fukamizu A., Hoshino T., Gotoh O., et al. (1991). Nucleotide sequence of the unique nitrate/nitrite-inducible cytochrome P-450 cDNA from Fusarium oxysporum. J. Biol. Chem. 266, 10632–10637. [PubMed] [Google Scholar]
- Klawonn I., Bonaglia S., Brüchert V., Ploug H. (2015). Aerobic and anaerobic nitrogen transformation processes in N2-fixing cyanobacterial aggregates. ISME J. 9, 1456–1466. 10.1038/ismej.2014.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Herman P. K., Emr S. D. (1990). The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54, 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver A. J., Donker H. J. L. (1926). Die Einheit in der Biochemie. Chem. Zelle Gewebe 13, 134–190. [Google Scholar]
- Kobayashi D. Y., Crouch J. A. (2009). Bacterial/fungal Interactions: from pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 47, 63–82. 10.1146/annurev-phyto-080508-081729 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Matsuo Y., Takimoto A., Suzuki S., Maruo F., Shoun H. (1996). Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271, 16263–16267. 10.1074/jbc.271.27.16263 [DOI] [PubMed] [Google Scholar]
- Koho K. A., Piña-Ochoa E., Geslin E., Risgaard-Petersen N. (2011). Vertical migration, nitrate uptake and denitrification: survival mechanisms of foraminifers (Globobulimina turgida) under low oxygen conditions. FEMS Microbiol. Ecol. 75, 273–283. 10.1111/j.1574-6941.2010.01010.x [DOI] [PubMed] [Google Scholar]
- Kraft B., Strous M., Tegetmeyer H. E. (2011). Microbial nitrate respiration - Genes, enzymes and environmental distribution. J. Biotechnol. 155, 104–117. 10.1016/j.jbiotec.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Kristensen E., Jensen M. H., Aller R. C. (1991). Direct measurement of dissolved inorganic nitrogen exchange and denitrification in individual polychaete (Nereis virens) burrows. J. Mar. Res. 49, 355–377. 10.1357/002224091784995855 [DOI] [Google Scholar]
- Larsen M., Thamdrup B., Shimmield T., Glud R. N. (2013). Benthic mineralization and solute exchange on a Celtic Sea sand-bank (Jones Bank). Prog. Oceanogr. 117, 64–75. 10.1016/j.pocean.2013.06.010 [DOI] [Google Scholar]
- Laughlin R., Stevens R. (2002). Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 66, 1540–1548. 10.2136/sssaj2002.1540 [DOI] [Google Scholar]
- Lomas M., Glibert P. (2000). Comparisons of nitrate uptake, storage, and reduction in marine diatoms and flagellates. J. Phycol. 36, 903–913. 10.1046/j.1529-8817.2000.99029.x [DOI] [Google Scholar]
- Lomas M. W., Glibert P. M. (1999). Temperature regulation of nitrate uptake: a novel hypothesis about nitrate uptake and reduction in cool-water diatoms. Limnol. Oceanogr. 44, 556–572. 10.4319/lo.1999.44.3.0556 [DOI] [Google Scholar]
- Lomstein E., Jensen M. H., Sørensen J. (1990). Intracellular and pools associated with deposited phytoplankton in a marine sediment (Aarhus Bight, Denmark). Mar. Ecol. Prog. Ser. 61, 97–105. 10.3354/meps061097 [DOI] [Google Scholar]
- Long A., Song B., Fridey K., Silva A. (2015). Detection and diversity of copper containing nitrite reductase genes (nirK) in prokaryotic and fungal communities of agricultural soils. FEMS Microbiol. Ecol. 91, 1–9. 10.1093/femsec/fiu004 [DOI] [PubMed] [Google Scholar]
- Lücking R., Huhndorf S., Pfister D. H., Plata E. R., Lumbsch H. T. (2009). Fungi evolved right on track. Mycologia 101, 810–822. 10.3852/09-016 [DOI] [PubMed] [Google Scholar]
- Lyons T. W., Reinhard C. T., Planavsky N. J. (2014). The rise of oxygen in Earth's early ocean and atmosphere. Nature 506, 307–315. 10.1038/nature13068 [DOI] [PubMed] [Google Scholar]
- Maeda K., Spor A., Edel-Hermann V., Heraud C., Breuil M. C., Bizouard F., et al. (2015). N2O production, a widespread trait in fungi. Sci. Rep. 5, 9697. 10.1038/srep09697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant H. K., Lavik G., Holtappels M., Kuypers M. M. (2014). The fate of nitrate in intertidal permeable sediments. PLoS ONE 9:e104517. 10.1371/journal.pone.0104517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Rotte C., Hoffmeister M., Theissen U., Gelius-Dietrich G., Ahr S., et al. (2003). Early cell evolution, eukaryotes, anoxia, sulfide, oxygen, fungi first (?), and a tree of genomes revisited. IUBMB Life 55, 193–204. 10.1080/1521654031000141231 [DOI] [PubMed] [Google Scholar]
- Martinoia E., Heck U., Wiemken A. (1981). Vacuoles as storage compartments for nitrate in barley leaves. Nature 289, 292–294. 10.1038/289292a0 [DOI] [Google Scholar]
- Matz M. V., Frank T. M., Marshall N. J., Widder E. A., Johnsen S. (2008). Giant deep-sea protist produces bilaterian-like traces. Curr. Biol. 18, 1849–1854. 10.1016/j.cub.2008.10.028 [DOI] [PubMed] [Google Scholar]
- McHatton S. C., Barry J. P., Jannasch H. W., Nelson D. C. (1996). High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62, 954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin L. K. (2011). A review of the evolution of the diatoms from the origin of the lineage to their populations, in The Diatom World, Cellular Origin, Life in Extreme Habitats and Astrobiology, Vol. 19, eds Seckbach J., Kociolek J. P. (Dordrecht; Heidelberg; London; New York, NY: Springer; ), 93–118. 10.1007/978-94-007-1327-7_4 [DOI] [Google Scholar]
- Mentel M., Martin W. (2008). Energy metabolism among eukaryotic anaerobes in light of Proterozoic ocean chemistry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2717–2729. 10.1098/rstb.2008.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothapo N., Chen H. H., Cubeta M. A., Grossman J. M., Fuller F., Shi W. (2015). Phylogenetic, taxonomic and functional diversity of fungal denitrifiers and associated N2O production efficacy. Soil Biol. Biochem. 83, 160–175. 10.1016/j.soilbio.2015.02.001 [DOI] [Google Scholar]
- Mothapo N. V., Chena H., Cubetab M. A., Shi W. (2013). Nitrous oxide producing activity of diverse fungi from distinct agroecosystems. Soil Biol. Biochem. 66, 94–101. 10.1016/j.soilbio.2013.07.004 [DOI] [Google Scholar]
- Müller M., Mentel M., van Hellemond J. J., Henze K., Woehle C., Gould S. B., et al. (2012). Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495. 10.1128/MMBR.05024-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mußmann M., Hu F. Z., Richter M., de Beer D., Preisler A., Jørgensen B. B., et al. (2007). Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 5:e230. 10.1371/journal.pbio.0050230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F. J., Machín F., Martín Y., Siverio J. M. (2006). Down-regulation of eukaryotic nitrate transporter by nitrogen-dependent ubiquitinylation. J. Biol. Chem. 281, 13268–13274. 10.1074/jbc.M601253200 [DOI] [PubMed] [Google Scholar]
- Needoba J. A., Harrison P. J. (2004). Influence of low light and a light: dark cycle on uptake, intracellular , and nitrogen isotope fractionation by marine phytoplankton. J. Phycol. 40, 505–516. 10.1111/j.1529-8817.2004.03171.x [DOI] [Google Scholar]
- Nielsen L. P. (1992). Denitrification in sediment determined from nitrogen isotope pairing. FEMS Microbiol. Lett. 86, 357–362. 10.1111/j.1574-6968.1992.tb04828.x [DOI] [Google Scholar]
- Nomaki H., Chikaraishi Y., Tsuchiya M., Toyofuku T., Ohkouchi N., Uematsu K., et al. (2014). Nitrate uptake by foraminifera and use in conjunction with endobionts under anoxic conditions. Limnol. Oceanogr. 59, 879–1888. 10.4319/lo.2014.59.6.1879 [DOI] [Google Scholar]
- Nomaki H., Chikaraishi Y., Tsuchiya M., Toyofuku T., Suga H., Sasaki Y., et al. (2015). Variation in the nitrogen isotopic composition of amino acids in benthic foraminifera: implications for their adaptation to oxygen-depleted environments. Limnol. Oceanogr. 60, 1906–1916. 10.1002/lno.10140 [DOI] [Google Scholar]
- Papaspyrou S., Smith C. J., Dong L. F., Whitby C., Dumbrell A. J., Nedwell D. B. (2014). Nitrate reduction functional genes and nitrate reduction potentials persist in deeper estuarine sediments. Why? PLoS ONE 9:e94111. 10.1371/journal.pone.0094111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parris D. J., Ganesh S., Edgcomb V. P., DeLong E. F., Stewart F. J. (2014). Microbial eukaryote diversity in the marine oxygen minimum zone off northern Chile. Front. Microbiol. 5:543. 10.3389/fmicb.2014.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J., Holzmann M., Berney C., Fahrni J., Gooday A. J., Cedhagen T., et al. (2003). The evolution of early Foraminifera. Proc. Natl. Acad. Sci. U.S.A. 100, 11494–11498. 10.1073/pnas.2035132100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña-Ochoa E., Høgslund S., Geslin E., Cedhagen T., Revsbech N. P., Nielsen L. P., et al. (2010a). Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc. Natl. Acad. Sci. U.S.A. 107, 1148–1153. 10.1073/pnas.0908440107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña-Ochoa E., Koho K. A., Geslin E., Risgaard-Petersen N. (2010b). Survival and life strategy of the foraminiferan Globobulimina turgida through nitrate storage and denitrification. Mar. Ecol. Prog. Ser. 417, 39–49. 10.3354/meps08805 [DOI] [Google Scholar]
- Ploug H., Kühl M., Buchholz-Cleven B., Jørgensen B. B. (1997). Anoxic aggregates - an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 13, 285–294. 10.3354/ame013285 [DOI] [Google Scholar]
- Prendergast-Miller M. T., Baggs E. M., Johnson D. (2011). Nitrous oxide production by the ectomycorrhizal fungi Paxillus involutus and Tylospora fibrillosa. FEMS Microbiol. Lett. 316, 31–35. 10.1111/j.1574-6968.2010.02187.x [DOI] [PubMed] [Google Scholar]
- Prieto M., Wedin M. (2013). Dating the diversification of the major lineages of Ascomycota (Fungi). PloS ONE 8:e65576. 10.1371/journal.pone.0065576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prihoda J., Tanaka A., de Paula W. B. M., Allen J. F., Tirichine L., Bowler C. (2012). Chloroplast-mitochondria cross-talk in diatoms. J. Exp. Bot. 63, 1543–1557. 10.1093/jxb/err441 [DOI] [PubMed] [Google Scholar]
- Prokopenko M. G., Sigman D. M., Berelson W. M., Hammond D. E., Barnett B., Chong L., et al. (2011). Denitrification in anoxic sediments supported by biological nitrate transport. Geochim. Cosmochim. Acta 75, 7180–7199. 10.1016/j.gca.2011.09.023 [DOI] [Google Scholar]
- Raimbault P., Mingazzini M. (1987). Diurnal variations of intracellular nitrate storage by marine diatoms: effects of nutritional state. J. Exp. Mar. Biol. Ecol. 112, 217–232. 10.1016/0022-0981(87)90070-0 [DOI] [Google Scholar]
- Revsbech N. P., Sorensen J., Blackburn T. H., Lomholt J. P. (1980). Distribution of oxygen in marine sediments measured with microelectrodes. Limnol. Oceanogr. 25, 403–411. 10.4319/lo.1980.25.3.0403 [DOI] [Google Scholar]
- Risgaard-Petersen N., Jensen K. (1997). Nitrification and denitrification in the rhizosphere of the aquatic macrophyte Lobelia dortmanna L. Limnol. Oceanogr. 42, 529–537. 10.4319/lo.1997.42.3.0529 [DOI] [Google Scholar]
- Risgaard-Petersen N., Langezaal A. M., Ingvardsen S., Schmid M. C., Jetten M. S. M., Op den Camp H. J. M., et al. (2006). Evidence for complete denitrification in a benthic foraminifer. Nature 443, 93–96. 10.1038/nature05070 [DOI] [PubMed] [Google Scholar]
- Rothe N., Gooday A. J., Cedhagen T., Hughes J. A. (2011). Biodiversity and distribution of the genus Gromia (Protista, Rhizaria) in the deep Weddell Sea (Southern Ocean). Polar Biol. 34, 69–81. 10.1007/s00300-010-0859-z [DOI] [Google Scholar]
- Satkoski A. M., Beukes N. J., Li W., Beard B. L., Johnson C. M. (2015). A redox-stratified ocean 3.2 billion years ago. Earth Planet Sci. Lett. 430, 43–53. 10.1016/j.epsl.2015.08.007 [DOI] [Google Scholar]
- Sayama M. (2001). Presence of nitrate-accumulating sulfur bacteria and their influence on nitrogen cycling in a shallow coastal marine sediment. Appl. Environ. Microbiol. 67, 3481–3487. 10.1128/AEM.67.8.3481-3487.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H. N., Brinkhoff T., Ferdelman T. G., Mariné M. H., Teske A., Jørgensen B. B. (1999). Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284, 493–495. 10.1126/science.284.5413.493 [DOI] [PubMed] [Google Scholar]
- Shoun H., Fushinobu S., Jiang L., Kim S.-W., Wakagi T. (2012). Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos. Trans. R. Soc. B 367, 1186–1194. 10.1098/rstb.2011.0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoun H., Tanimoto T. (1991). Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266, 11078–11082. [PubMed] [Google Scholar]
- Sierra R., Matz M. V., Aglyamova G., Pillet L., Decelle J., Not F., et al. (2013). Deep relationships of Rhizaria revealed by phylogenomics: a farewell to Haeckel's Radiolaria. Mol. Phylogenet. Evol. 67, 53–59. 10.1016/j.ympev.2012.12.011 [DOI] [PubMed] [Google Scholar]
- Sims P. A., Mann D. G., Medlin L. K. (2006). Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402. 10.2216/05-22.1 [DOI] [Google Scholar]
- Steward C. C., Nold S. C., Ringelberg D. B., White D. C., Lovell C. R. (1996). Microbial biomass and community structures in the burrows of bromophenol producing and non-producing marine worms and surrounding sediments. Mar. Ecol. Prog. Ser. 133, 149–165. 10.3354/meps133149 [DOI] [Google Scholar]
- Stief P., de Beer D. (2006). Probing the microenvironment of freshwater sediment macrofauna: implications of deposit-feeding and bioirrigation for nitrogen cycling. Limnol. Oceanogr. 51, 2538–2548. 10.4319/lo.2006.51.6.2538 [DOI] [Google Scholar]
- Stief P., Fuchs-Ocklenburg S., Kamp A., Manohar C. S., Houbraken J., Boekhout T., et al. (2014). Dissimilatory nitrate reduction by Aspergillus terreus isolated from the seasonal oxygen minimum zone in the Arabian Sea. BMC Microbiol. 14:35. 10.1186/1471-2180-14-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief P., Kamp A., de Beer D. (2013). Role of diatoms in the spatial-temporal distribution of intracellular nitrate in intertidal sediment. PLoS ONE 8:e73257. 10.1371/journal.pone.0073257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T., Behnke A., Christen R., Amaral-Zettler L., Rodriguez-Mora M. J., Chistoserdov A., et al. (2009). Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 7:72. 10.1186/1741-7007-7-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohm T. O., Griffin B., Zumft W. G., Schink B. (2007). Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 73, 1420–1424. 10.1128/AEM.02508-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweerts J.-P. R. A., de Beer D. (1989). Microelectrode measurements of nitrate gradients in the littoral and profundal sediments of a meso-eutrophic lake (Lake Vechten, The Netherlands). Appl. Environ. Microbiol. 55, 754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki K., Shoun H., Yamaguchi M., Takeo K., Nakamura A., Hoshino T., et al. (2004). Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol - Role of acetyl CoA synthetase in anaerobic ATP synthesis. J. Biol. Chem. 279, 12414–12420. 10.1074/jbc.M313761200 [DOI] [PubMed] [Google Scholar]
- Takaya N. (2009). Response to hypoxia, reduction of electron acceptors, and subsequent survival by filamentous fungi. Biosci. Biotechnol. Biochem. 73, 1–8. 10.1271/bbb.80487 [DOI] [PubMed] [Google Scholar]
- Takaya N., Suzuki S., Kuwazaki S., Shoun H., Maruo F., Yamaguchi M., et al. (1999). Cytochrome p450nor, a novel class of mitochondrial cytochrome P450 involved in nitrate respiration in the fungus Fusarium oxysporum. Arch. Biochem. Biophys. 372, 340–346. 10.1006/abbi.1999.1499 [DOI] [PubMed] [Google Scholar]
- Tantanasarit C., Englande A., Babel S. (2013). Nitrogen, phosphorus and silicon uptake kinetics by marine diatom Chaetoceros calcitrans under high nutrient concentrations. J. Exp. Mar. Biol. Ecol. 446, 67–75. 10.1016/j.jembe.2013.05.004 [DOI] [Google Scholar]
- Thamdrup B. (2012). New Pathways and processes in the global nitrogen cycle. Annu. Rev. Ecol. Evol. Syst. 43, 407–428. 10.1146/annurev-ecolsys-102710-145048 [DOI] [Google Scholar]
- Thornton D. C. O. (2002). Diatom aggregation in the sea: mechanisms and ecological implications. Eur. J. Phycol. 37, 149–161. 10.1017/S0967026202003657 [DOI] [Google Scholar]
- Tsuruta S., Takaya N., Zhang L., Shoun H., Kimura K., Hamamoto M., et al. (1998). Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 168, 105–110. 10.1111/j.1574-6968.1998.tb13262.x [DOI] [PubMed] [Google Scholar]
- Unkles S. E., Wang R. C., Wang Y., Glass A. D. M., Crawford N. M., Kinghorn J. R. (2004). Nitrate reductase activity is required for nitrate uptake into fungal but not plant cells. J. Biol. Chem. 279, 28182–28186. 10.1074/jbc.M403974200 [DOI] [PubMed] [Google Scholar]
- Usuda K., Toritsuka N., Matsuo Y., Kim D. H., Shoun H. (1995). Denitrification by the fungus Cylindrocarpon tonkinense: anaerobic cell growth and two isozyme forms of cytochrome P-450nor. Appl. Environ. Microbiol. 61, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M. (2011). Mitochondria and the rise of eukaryotes. Bioscience 61, 594–601. 10.1525/bio.2011.61.8.5 [DOI] [Google Scholar]
- Villareal T. A., Altabet M. A., Culverrymsza K. (1993). Nitrogen transport by vertically migrating diatom mats in the North Pacific Ocean. Nature 363, 709–712. 10.1038/363709a0 [DOI] [Google Scholar]
- Villareal T. A., Joseph L., Brzezinski M. A., Shipe R. F., Lipschultz F., Altabet M. A. (1999). Biological and chemical characteristics of the giant diatom Ethmodiscus (Bacillariophyceae) in the central north pacific gyre. J. Phycol. 35, 896–902. 10.1046/j.1529-8817.1999.3550896.x [DOI] [Google Scholar]
- Wei W., Isobe K., Shiratori Y., Nishizawa T., Ohte N., Ise Y., et al. (2015). Development of PCR primers targeting fungal nirK to study fungal denitrification in the environment. Soil Biol. Biochem. 81, 282–286. 10.1016/j.soilbio.2014.11.026 [DOI] [Google Scholar]
- Wenzhöfer F., Glud R. N. (2004). Small-scale spatial and temporal variability in coastal benthic O2 dynamics: effects of fauna activity. Limnol. Oceanogr. 49, 1471–1481. 10.4319/lo.2004.49.5.1471 [DOI] [Google Scholar]
- Williams K. P., Sobral B. W., Dickerman A. W. (2007). A robust species tree for the Alphaproteobacteria. J. Bacteriol. 189, 4578–4586. 10.1128/JB.00269-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden A. Z., Follows M. J., Giovannoni S. J., Wilken S., Zimmerman A. E., Keeling P. J. (2015). Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 735. 10.1126/science.1257594 [DOI] [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G. J., Woese C. R. (1985). Mitochondrial origins. Proc. Natl. Acad. Sci. U.S.A. 82, 4443–4447. 10.1073/pnas.82.13.4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Takaya N., Nakamura A., Yamaguchi M., Takeo K., Shoun H. (2002). Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J. Biol. Chem. 277, 1892–1896. 10.1074/jbc.M109096200 [DOI] [PubMed] [Google Scholar]
- Zumft W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.