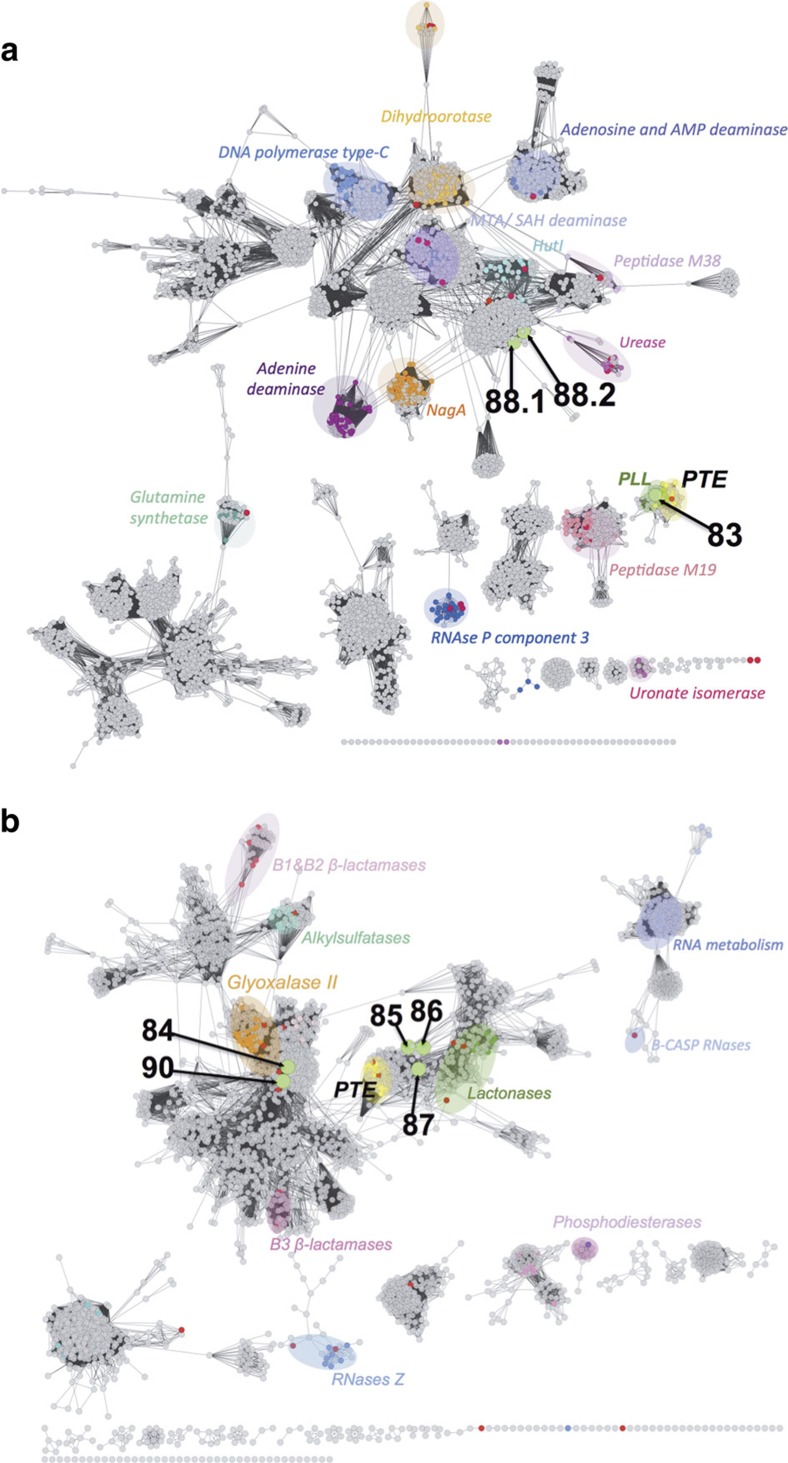
Figure 4. SSNs highlight the novelty of the triesterases hits.
Hits are spread over (a) the AH clan (Pfam number: CL0034) and (b) the MBL superfamily (Pfam family PF00753). Bright green nodes represent the sequences of metagenomic hits identified in this work. Annotations retrieved from the Uniprot database were used to putatively characterize sequence clusters (represented by coloured nodes). To confirm these annotations, experimentally characterized proteins (red nodes) were added into each network. Previously described OP-degrading enzymes (PTE, phosphotriesterase) are highlighted in yellow; PLL (PTE-like lactonases) are reported in the AH superfamily. 5,042 and 2,984 sequences define the AH and the MBL networks, respectively. Edges lengths represent protein sequence similarity. Only edges corresponding to similarity scores below E-values of 1 × e−9 (AH) and 1 × e−14 (MBL) are considered; the worst edges displayed correspond to a median 26.5% (or 27.9%) identity over an alignment length of 210 (or 218) residues for the AH (and MBL) networks, respectively. See also the position of additional hits in the α/β superfamily (Supplementary Fig. 15).