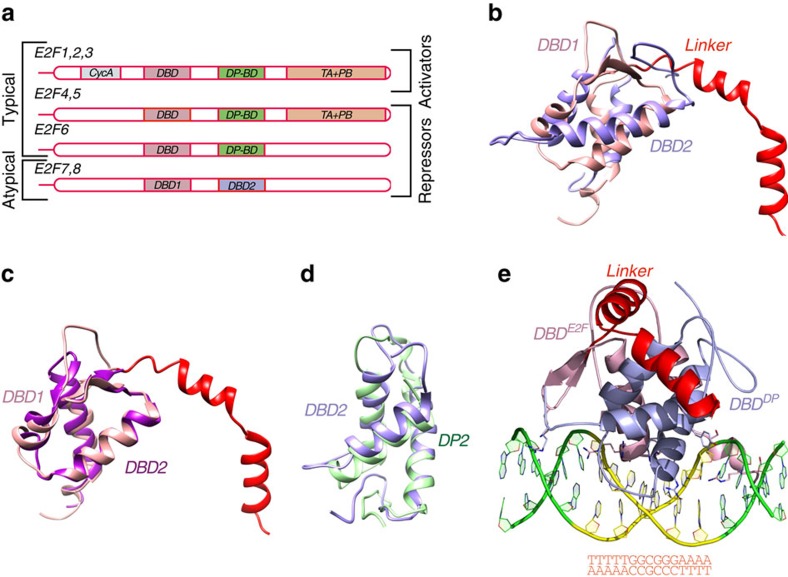
Figure 1. Structure of E2F8.
(a) Schematic representation of the structural organization of E2F transcription factors. Key: CycA: cyclin A-binding domain; DBD: DNA-binding domain; DP-BD: DP-binding domain; TA+PB: transactivation and pocket protein-binding domains. Note that the typical E2Fs have DP-binding domains, which are replaced by a second DBD in the atypical E2Fs. (b) Superimposition of E2F8 DBD1 (pink) and DBD2 (blue) (r.m.s.d.=7.8 Å); the linker between the two DBDs is in red. (c,d) Superimpositions of E2F8 DBD1 (pink) to E2F4 (magenta) (r.m.s.d.=1.36 Å; PDB ID 1CF7) and E2F8 DBD2 (blue) to DP2 (green) (r.m.s.d.=1.9 Å; PDB ID 1CF7). The 23 amino acids of the linker close to DBD1 are folded into two α-helices, whereas the remaining 53 amino acids connected to DBD2 are disordered. Note the high similarity between the domains. (e) Structure of the E2F8 protein containing DBD1 (DBDE2F, pink) and DBD2 (DBDDP, blue) bound to a 15-base pair DNA fragment (green and yellow). Residues responsible for the motif recognition are presented as ball-and-stick models and coloured by atom (carbon: chain colour; nitrogen: blue; oxygen: red). The sequence of the DNA fragment is also shown.