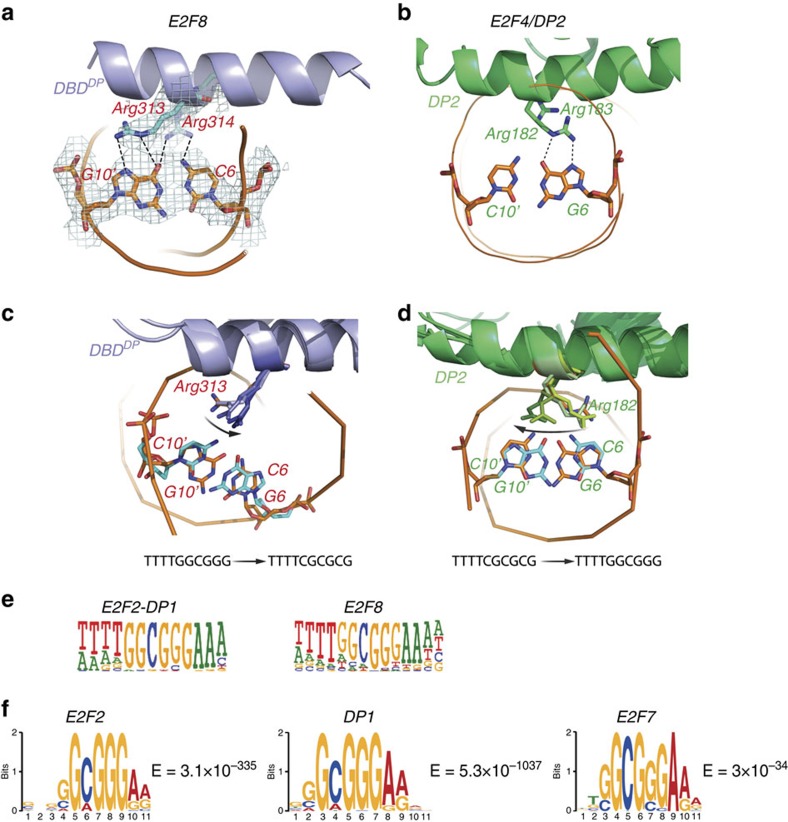
Figure 3. Atypical and typical E2Fs prefer similar motifs.
(a) Close-up view of the contacts between the E2F8 DBDDP arginines 313 and 314 and the DNA base pair G10′-C6 (C opposite to the capitalized G in tggcgGga). (b) The corresponding contact between DP2 and DNA in the E2F4/DP2-DNA complex. Note that the bound DNA sequence is different, and the arginine 182 of DP2 makes contact to a guanine on the opposite strand of DNA compared to that recognized by the corresponding Arg313 of E2F8. (c,d) Molecular dynamics simulations of the E2F8 DBDDP bound to 5′-TTTTTCGCGCG-3′ (c) and the DP2 protein bound to 5′-TTTTGGCGGG-3′ (d). Five snapshots taken every 20 ns are shown. The original position of the Arg residue and the original base pair are coloured in orange. All following positions are coloured in progressively darkening color. The mutated base pairs are coloured in blue. Note that upon change of the underlying DNA sequence, the arginine moves (arrow) to the position observed in the other crystal, suggesting that atypical and typical E2Fs can recognize the same set of sequences. (e) HT-SELEX analyses for E2F2/DP1 complex and E2F8 performed in this study reveal that a typical E2F/DP and an atypical E2F prefer sequences that are very similar to each other (note that the obtained E2F8 motif is very similar to that reported in ref. 17). (f) Typical and atypical E2Fs prefer similar sequences in vivo. The most enriched motifs from genomic sequences bound by E2F2 and DP1 in ChIP-exo experiments performed in this work are virtually identical to a motif that is enriched by E2F7 in ChIP-seq (data from ref. 44). MEME E-values of the motifs are also shown.