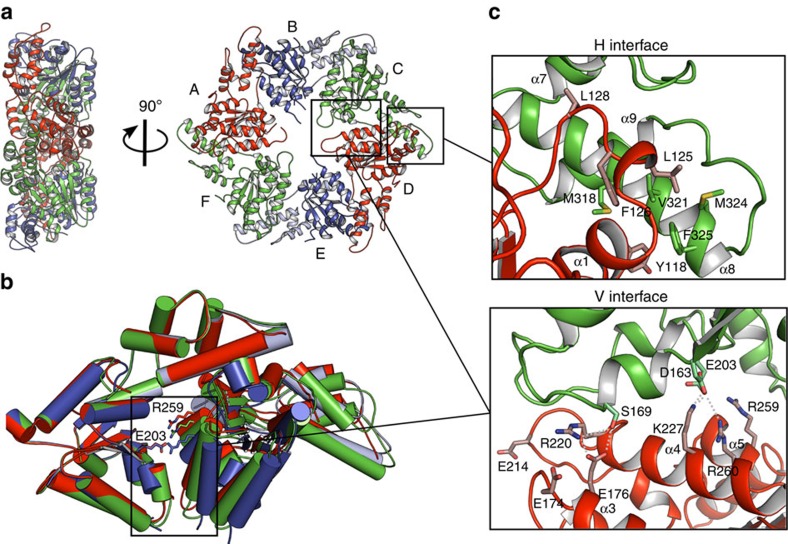
Figure 2. Crystal structure of the MsVps4ΔMIT pseudohexamer.
(a) Cartoon representation of the MsVps4ΔMIT pseudohexamer viewed from two orientations. Asymmetric MsVps4ΔMIT has twofold non-crystallographic symmetry with opposing protomers making identical interactions. The protomers are labelled from A to F and identical protomers are represented in the same colour, except for protomers B and E, whose small ATPase domain is shown in light blue and the large ATPase domain in dark blue. (b) Superposition of the three different dimers present in the pseudohexamer, formed by A–B, B–C and C–D. The dimeric substructures are superimposed by aligning the large ATPase domain of the first protomer. This superposition reveals different positions of the large ATPase domain. The catalytic Glu203 (left subunit) and the arginine finger residue Arg259 (right subunit) are represented with sticks. (c) Molecular interactions at the H interface of the C–D dimer and molecular interactions at the variable V interface of the same dimer. A list of the molecular interactions of the three interfaces is shown in Supplementary Table 1.