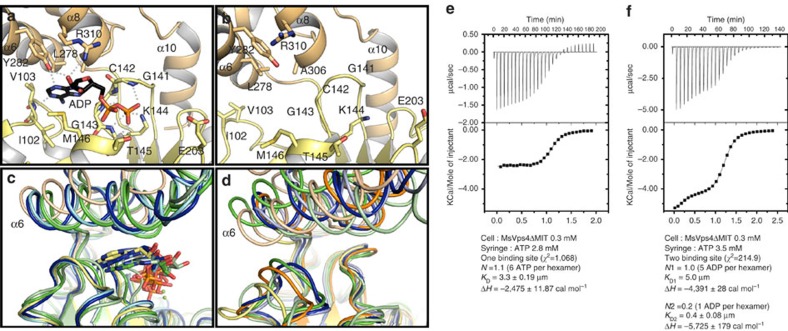
Figure 3. Crystal structure of MsVps4ΔL-MIT bound to ADP.
View of the nucleotide-binding site. (a) Structure of ADP bound MsVps4ΔL-MIT. The MsVps4ΔL-MIT large ATPase domain (yellow) and small ATPase domain (beige) are represented in cartoon. ADP and interacting residues are shown as sticks and coloured by atom type. Polar interactions are indicated by grey dashes. (b) Close-up of the nucleotide-binding site of apo MsVps4ΔMIT (protomer F). (c) Superposition of the ADP-bound MsVps4ΔL-MIT (yellow and beige), ATPγS-bound yeast Vps4 (green, pdb 3EIH), ADP-bound yeast Vps4 (pale green, pdb 2QPA), ATP-bound mouse VPS4B (blue, pdb 2ZAN) and ADP-bound mouse VPS4B (pale blue, pdb 2ZAO). (d) Superposition of different Vps4 structures with no nucleotide bound. Superposition of apo MsVps4ΔMIT (yellow and beige), apo SsoVps4 (orange, pdb 4LGM, Cl− bound in the P-loop), yeast Vps4p (green, pdb 3EIH, molecule C, ethylene glycol-Mg2+ bound in the P-loop), apo yeast Vps4p (pale green, pdb 2RKO), hVPS4B (blue, pdb 1XWI, SO4 bound in the P-loop) and mouse VPS4B (pale blue, pdb 2ZAM). The structures are superimposed by aligning the large ATPase domains. Nucleotides are represented in sticks in the same colour as the corresponding structures. (e) ITC analyses of ATP binding to MsVps4ΔMIT. ITC data were recorded on successive injection of ATP into the cell containing 300 μM MsVps4ΔMIT, which permits nucleotide-independent hexamer formation. This revealed a 1:1 binding mode for ATP. (f) ITC analyses of ADP binding to MsVps4ΔMIT at a concentration of 300 μM. The experimental data fit best to a two binding-site model with one high-affinity binding site and five lower-affinity binding sites. Experiments shown in e and f were repeated three times.