Figure 5. Proteomics and cytotoxicity analysis of n-butylamine bioconjugated gluten.
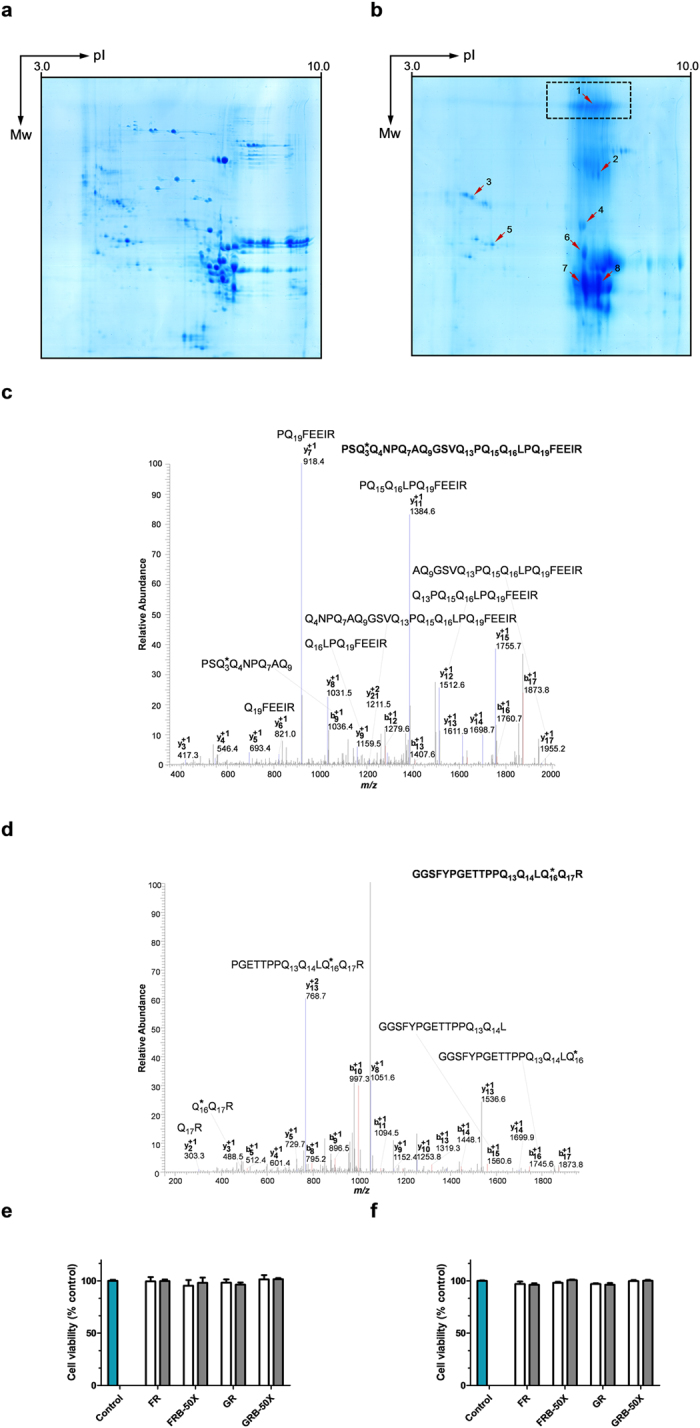
Two-dimensional electrophoresis pattern (IEF × SDS-PAGE) of the gluten, original (a) and derivatised with n-butylamine (b) as amine nucleophile under reducing conditions. The arrowheads point to the excised and identified protein spots (please see Table 1, for spot identification). The dashed rectangle indicates protein aggregates. Tandem mass (MS/MS) spectra of (c) α-/β-gliadin derived tryptic digest peptide [precursor ion at m/z (+2) = 1396.2] from spot 7/8, excised from 2-DE gel of GRB 50X; (d) HMW-GS derived tryptic digest peptide [precursor ion at m/z (+2) = 1024.5] from spot 1, excised from 2-DE gel of GRB 50X. * indicates the modified glutamine residue. Alamar Blue (AB) assay for cell viability quantification using Caco-2 cells, after 24 h (e) or 48 h (f) treatment with different concentrations of peptic-tryptic digests of wheat flour and gluten, original and derivatised with n-butylamine, as amine nucleophile, under reducing conditions. For sample nomenclature please consult Fig. 6. A total of n = 3 different experiments (each with four replicates), from different cell passages. Error bars represent the standard deviation Empty bars ( ) denote 0.5 mg/mL; and filled bars (
) denote 0.5 mg/mL; and filled bars ( ) denote 1.0 mg/mL.
) denote 1.0 mg/mL.
