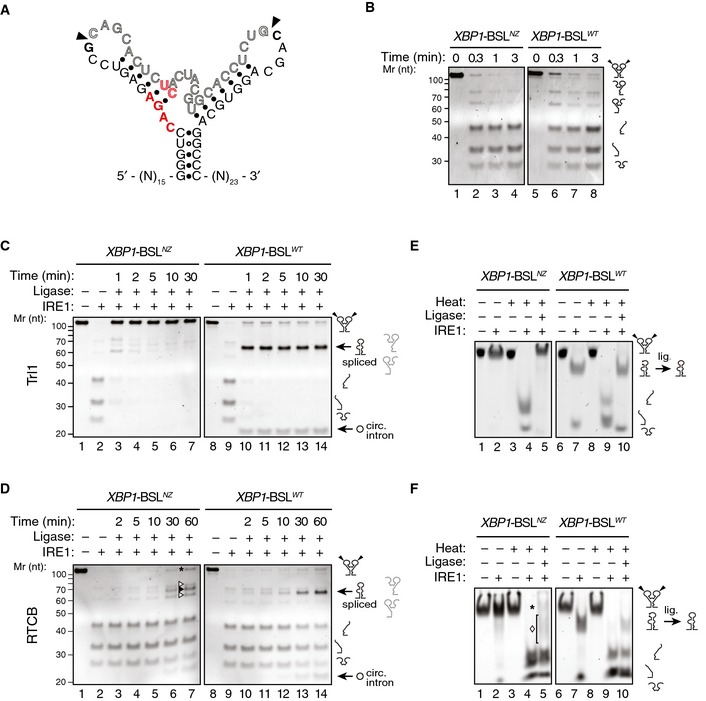
Figure 4. Formation of ES1 is required for XBP1 mRNA splicing.
-
ASecondary structure of the mutant XBP1‐ BSLNZ RNA transcript. The base substitutions indicated in red disrupt exon–exon base pairing. Arrowheads: scissile bonds.
-
BTBE–urea–PAGE gels showing cleavage of the XBP1‐ BSLNZ or XBP1‐ BSLWT RNAs by IRE1. The RNAs were incubated with 0.5 μM of IRE1α‐KR43 for the indicated times.
-
C, DTBE–urea–PAGE gels showing IRE1‐mediated cleavage of the XBP1‐ BSLWT or XBP1‐ BSLNZ RNAs and their splicing after incubation of the cleaved RNAs with Trl1 (C), or the RTCB complex (D). All reactions with RTCB were supplemented with recombinant archease. Circ. intron: circularized intron. Asterisk: input RNA (both exons religated to intron). Open arrowheads: incomplete splice products (5′ or 3′ exons ligated to the intron). Closed arrowhead: spliced product. The RNAs in (C, D) were incubated with 0.5 μM of IRE1α‐KR43 for cleavage and with the depicted tRNA ligase for the indicated times. The light grey pictograms on the right of the gels indicate the partially cleaved or incompletely spliced RNAs (containing a single exon and the intron).
-
E, FNative PAGE gels showing IRE1‐mediated cleavage of the XBP1‐ BSLWT or XBP1‐ BSLNZ RNAs and their splicing after subsequent incubation of the cleaved RNAs with Trl1 (E) or the RTCB complex (F). The RNAs in (E, F) were incubated with 0.5 μM of IRE1α‐KR43 prior to thermal denaturation and ligation. The ligation reactions in (F) were supplemented with recombinant archease. Asterisk in (F): input RNA (both exons re‐ligated to intron). Diamond in (F): incompletely spliced products (a single exon ligated to the intron). Note that the spliced product (exon–exon ligation) co‐migrates with the incompletely spliced products, which makes it indistinguishable in native PAGE conditions.
