Figure 8.
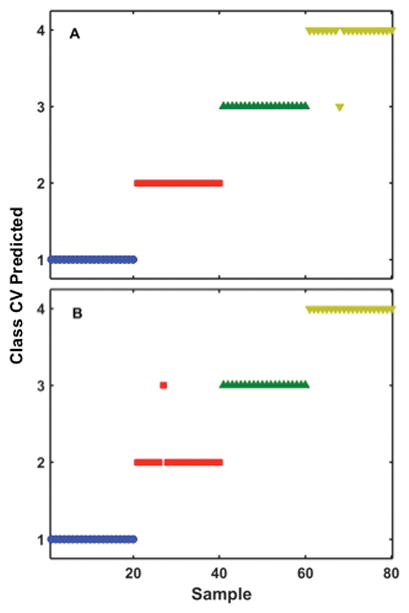
SVM-DA cross-validation (CV) predicted plots for (A) high, and (B) low NA-EIV assays. A total of 80 spectra are represented in this plot, with 20 replicates in each group. Each colored symbol represents the SVM CV predicted class membership for an individual SERS spectrum after incubation of synthetic RNA strains at 37°C for 2 hours with DNA probe. The colored symbol and sample numbers are identical to the samples described in Figures 6 and 7.
(A) The colored symbols represents SERS spectra containing: the high NA-EIV RNA strains of A/ck/Hubei/327/2004/H5N1 (
 ), and A/WSN/33/H1N1 (
), and A/WSN/33/H1N1 (
 ) and the low NA-EIV RNA strains of A/Gs/Gd/1/96/H5N1 (
) and the low NA-EIV RNA strains of A/Gs/Gd/1/96/H5N1 (
 ) incubated with high NA-EIV DNA probe. High DNA probe with MCH spacer was used as a control (
) incubated with high NA-EIV DNA probe. High DNA probe with MCH spacer was used as a control (
 ).
).
(B) The colored symbols represents SERS spectra containing: the low NA-EIV RNA strains of A/Gs/Gd/1/96/H5N1 (
 ) and the high NA-EIV RNA strains of A/ck/Hubei/327/2004/H5N1 (
) and the high NA-EIV RNA strains of A/ck/Hubei/327/2004/H5N1 (
 ), and A/WSN/33/H1N1 (
), and A/WSN/33/H1N1 (
 ) incubated with low NA-EIV DNA probe. Low NA-EIV DNA probe with MCH spacer was used as control (
) incubated with low NA-EIV DNA probe. Low NA-EIV DNA probe with MCH spacer was used as control (
 ).
).
