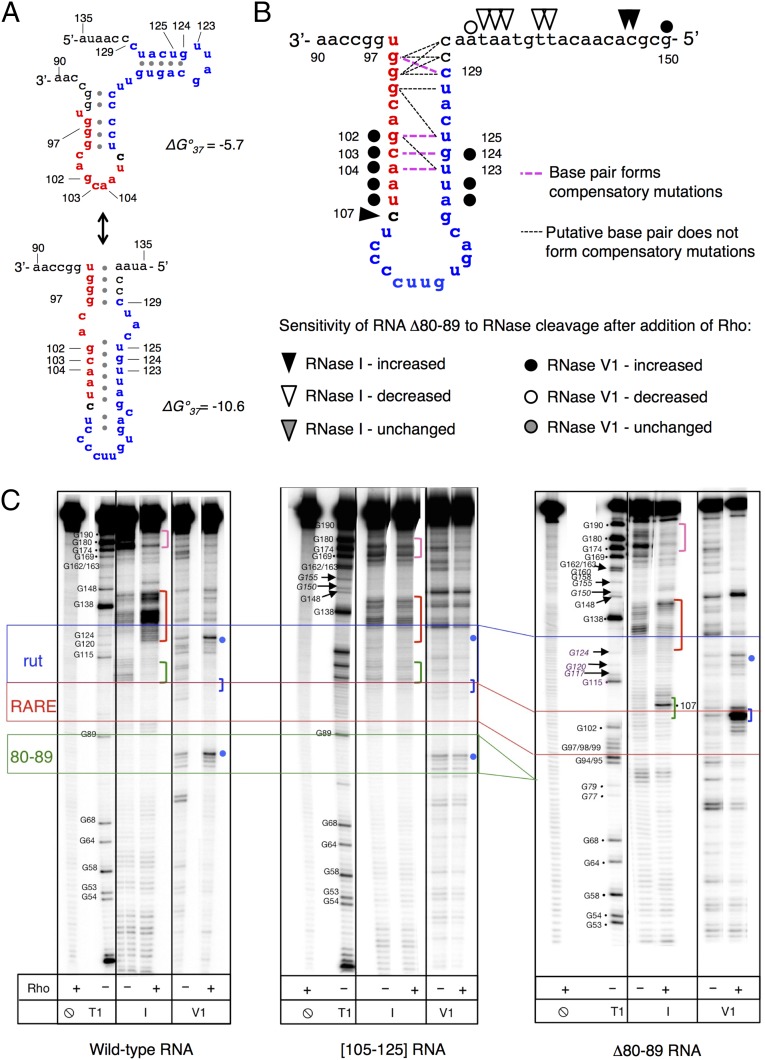
Fig. 4.
RARE promotes the formation of an unusual complex between the mgtCBR leader RNA and Rho. (A) Putative secondary structures formed by RARE (red) and rut (blue) in the mgtCBR leader. Structures were generated using the Mfold web server for nucleic acid folding and hybridization prediction (46). (B) Graphic representation of the proposed structure formed by RARE (red) and the rut (blue) in the mgtCBR leader based on RARE mutagenesis (Fig. 3C), compensatory mutation analysis (Fig. S4A), and the structural probing shown below. Mutant combinations that restored fluorescence to the levels displayed by the Δ80–89 variant are shown in bold pink dashed lines; mutant combinations that were tested but did not restore Δ80–89 function are shown in thin black dashed lines. See text for details. (C) Enzymatic probing of the wild-type mgtCBR leader RNA and the [105–125] and Δ80–89 variants. The 5′ 32P-labeled fragment of the mgtCBR leader RNA was treated with different RNases in the presence or absence of Rho. T1 is for RNase T1 (cuts at G residues, used as a molecular marker), I is for RNase I, and V1 is for RNase V1. Positions of interest are marked with colored dots and brackets (see text). Colored lines show borders of relevant elements: blue for rut, red for RARE, and green for the left arm of stem-loop B (80–89). Only the relevant lanes from a representative gel are shown. The experiment was performed at least three times for each sample.