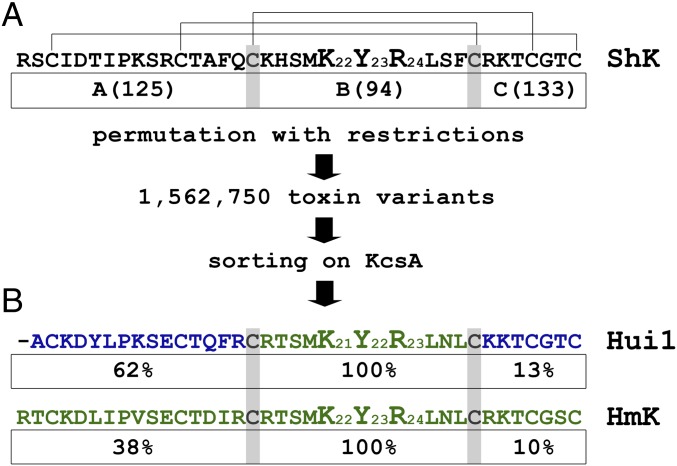
Fig. 1.
Library design and phage sorting. KcsA channels select phage-expressing Hui1 and HmK by binding. Phage preparation, library construction and sorting, and synthesis of KcsA and toxins are described in Materials and Methods. Single-letter codes for amino acids are standard. (A) SAK1 sequences (150) were aligned on conserved Cys residues (indicated by the gray highlighting) to define three domains that correspond to ShK residues Arg1-Gln16, Lys18-Phe27, and Arg29-Cys35, respectively. This yields 125, 94, and 133 unique A, B, and C domains, respectively (Table S1), which combine to produce ∼1.5 million ABC peptides. After five rounds of selection on KcsA, phage enrichment was observed compared with the NeutrAvidin (nAv) control (Fig. S1). (B) Hui1, a novel toxin isolated from the library, is composed of residues present in two parental toxins, AETX-K (blue) and HmK (green). Under each of the three domains in Hui1 is indicated its frequency of representation (in percent) among the 10 peptides enriched by phage selection on KcsA (Table S2).