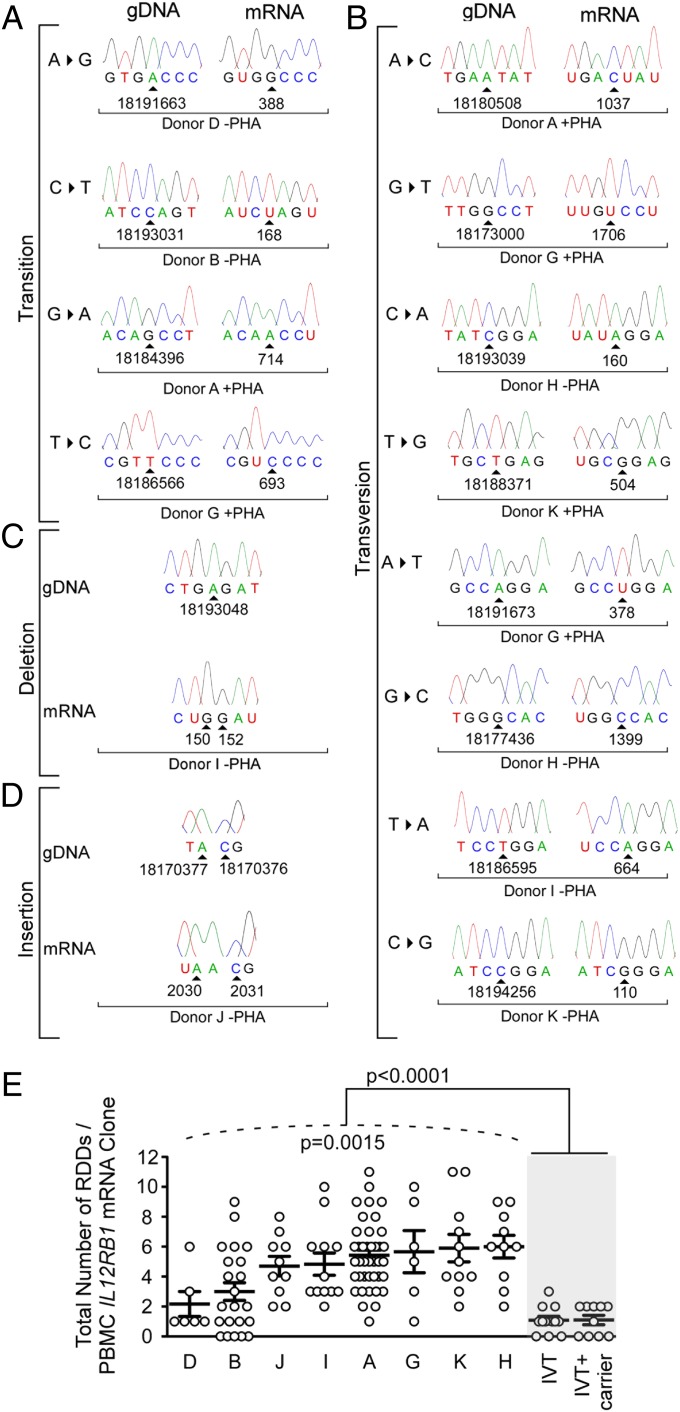
Fig. 2.
The extent of IL12RB1 RDD introduction varies significantly between individuals. After their purification from the PBMCs of eight different donors (A, B, D, and G–K), IL12RB1 mRNAs were amplified and cloned into the sequencing vector pUC19. Individual clones from each PBMC IL12RB1 library were randomly chosen for Sanger sequencing (two to three times coverage; average of 15 mRNA clones per donor). (A–D) The IL12RB1 RDDs detected by Sanger sequencing comprised multiple RDD types, including (A) transitions, (B) transversions, (C) deletions, and (D) insertions. Shown for each RDD type in (A and B) are sequencing chromatograms of the IL12RB1 gDNA sequence (Left) next to the corresponding IL12RB1 mRNA clone sequence from the same donor (Right). (C and D) The sequence of a donor gDNA (Top) and the corresponding mRNA clone (Bottom) sequence. (A–D) The genomic location of the deoxyribonucleotide encoding the affected ribonucleotide is indicated at the bottom of each sequencing trace, as is the ribonucleotide number and associated mRNA clone information. (E) The extent to which RDDs occurred in IL12RB1 mRNA clones expressed by her or his PBMCs (PHA− and PHA+ data are combined; Dataset S4). Each circle represents a single IL12RB1 mRNA clone and its IL12RB1 RDD frequency (i.e., the number of RDDs present divided by the mRNA clone length). The extent to which donors’ individual RDD frequency data significantly differed from one another is indicated below the hatched line (P = 0.0015, as measured using a mixed-effects Poisson regression model); the extent to which the RDD frequency data from all donors differed from recombinant polymerase error rate (in vitro transcription), which was determined both in the presence and absence of nonspecific carrier DNA, is indicated at the top of the graph (P < 0.0001, using Student’s t test).